In this post, we walk you through the steps to build machine learning (ML) models in Amazon SageMaker with data stored in Amazon HealthLake using two example predictive disease models we trained on sample data using the MIMIC-III dataset. This dataset was developed by the MIT lab for Computational Physiology and consists of de-identified healthcare data associated with approximately 60,000 ICU admissions. The dataset includes multiple attributes about the patients like their demographics, vital signs, and medications, along with their clinical notes. We first developed the models using the structured data such as demographics, vital signs, and medications. Then we augmented these models with additional data extracted and normalized from clinical notes to test and compare their performance. In both these experiments, we found an improvement in model performance when modelled as a supervised learning (classification) or an unsupervised learning (clustering) problem. We present our findings and the setup of the experiments in this post.
Why multiple modalities?
Modality can be defined as the classification of a single independent sensory input/output between a computer and a human. For example, we can see objects and hear sounds by using our senses. These can be considered as two separate modalities. Datasets that represent multiple modalities are categorized as a multi-modal dataset. For instance, images can consist of tags that help search and organize them, and textual data can contain images to explain what’s in the image. When medical practitioners make clinical decisions, it’s usually based on information gathered from a variety of healthcare data modalities. A physician looks at patient’s observations, their past history, their scans, and even physical characteristics of the patient during the visit to make a definitive diagnosis. ML models need to take this into account when trying to achieve real-world performance. The post Building a medical image search platform on AWS shows how you can combine features from medical images and their corresponding radiology reports to create a medical image search platform. The challenge with creating such models is the preprocessing of these multi-modal datasets and extracting appropriate features from them.
Amazon HealthLake makes it easier to train models on multi-modal data
Amazon HealthLake is a HIPAA eligible service that enables healthcare providers, health insurance companies, and pharmaceutical companies to store, transform, query, and analyze health data on the AWS Cloud at petabyte scale. As part of the transformation, Amazon HealthLake tags and indexes unstructured data using specialized ML models. These tags and indexes can be used to query and search as well as understand relationships in the data for analytics.
When you export data from Amazon HealthLake, it adds a resource called DocumentReference to the output. This resource consists of clinical entities (Like medications, medical conditions, anatomy, and Protected Health Information (PHI)), the RxNorm codes for medications, and the ICD10 codes for medical conditions that are automatically derived from the unstructured notes about the patients. These are additional attributes about the patients that are embedded within the unstructured portions of their clinical records and would have been largely ignored for downstream analysis. Combining the structured data from the EHR with these attributes provides a more holistic picture of the patient and their conditions. To help determine the value of these attributes, we created a couple of experiments around clinical outcome prediction.
Architecture overview
The following diagram illustrates the architecture for our experiments.
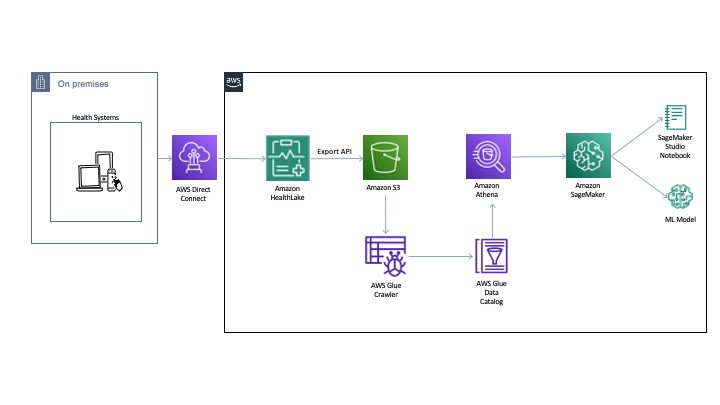
You can export the normalized data to an Amazon Simple Storage Service (Amazon S3) bucket using the Export API. Then we use AWS Glue to crawl and build a catalog of the data. This catalog is shared by Amazon Athena to run the queries directly off of the exported data from Colossus. Athena also normalizes the JSON format files to rows and columns for easy querying. The DocumentReference resource JSON file is processed separately to extract indexed data derived from the unstructured portions of the patient records. The file consists of an extension tag that has a hierarchical JSON output consisting of patient attributes. There are multiple ways to process this file (like using Python-based JSON parsers or even string-based regex and pattern matching). For an example implementation, see the section Connecting Athena with HealthLake in the post Population health applications with Amazon HealthLake – Part 1: Analytics and monitoring using Amazon QuickSight.
Example setup
Accessing the MIMIC-III dataset requires you to request access. As part of this post, we don’t distribute any data but instead provide the setup steps so you can replicate these experiments when you have access to MIMIC-III. We also publish our conclusions and findings from the results.
For the first experiment, we build a binary disease classification model to predict patients with congestive heart failure (CHF). We measure its performance using accuracy, ROC, and confusion matrix for both structured and unstructured patient records. For the second experiment, we cluster a cohort of patients into a fixed number of groups and visualize the cluster separation before and after the addition of the unstructured patient records. For both our experiments, we build a baseline model and compare it with the multi-modal model, where we combine existing structured data with additional features (ICD-10 codes and Rx-Norm codes) in our training set.
These experiments are not intended to produce a state-of-the-art model on real-world datasets; its purpose is to demonstrate how you can utilize features exported from Amazon Healthlake for training models on structured and unstructured patient records to improve your overall model performance.
Features and data normalization
We took a variety of features related to patient encounters to train our models. This included the patient demographics (gender, marital status), the clinical conditions, procedures, medications, and observations. Because each patient could have multiple encounters consisting of multiple observations, clinical conditions, procedures, and medications, we normalized the data and converted each of these features into a list. This allowed us to get a training set with all these features (as a list) for each patient.
Similarly, for the unstructured features that Amazon Healthlake converted into the DocumentReference resource, we extracted the ICD-10 codes and Rx-Norm codes (using the methods described in the architecture) and converted them into feature vectors.
Feature engineering and model
For the categorical attributes in our dataset, we used a label encoder to convert the attributes into a numerical representation. For all other list attributes, we used term frequency-inverse document frequency (FI-IDF) vectors. This high-dimensional dataset was then shuffled and divided into 80% train and 20% test sets for training and evaluation of the models, respectively. For training our model, we used the gradient boosting library XGBoost. We considered mostly default hyperparameters and didn’t perform any hyperparameter tuning, because our objective was only to train a baseline model with structured patient records and then show improvement on those results with the unstructured features. Adopting better hyperparameters or changing to other feature engineering and modelling approaches can likely improve these results.
Example 1: Predicting patients with a congestive heart failure
For the first experiment, we took 500 patients with a positive CHF diagnosis. For the negative class, we randomly selected 500 patients who didn’t have a CHF diagnosis. We removed the clinical conditions from the positive class of patients that were directly related to CHF. For example, all the patients in the positive class were expected to have ICD-9 code 428, which stands for CHF. We filtered that out from the positive class to make sure the model is not overfitting on the clinical condition.
Baseline model
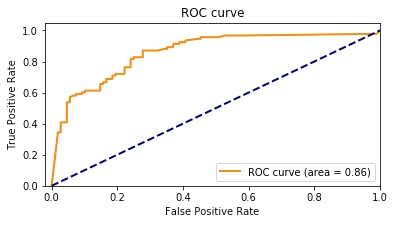
Our baseline model had an accuracy of 85.8%. The following graph shows the ROC curve.
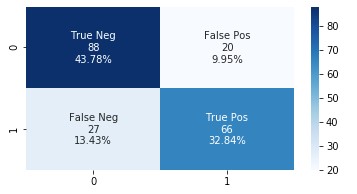
The following graph shows the confusion matrix.
Amazon HealthLake augmented model
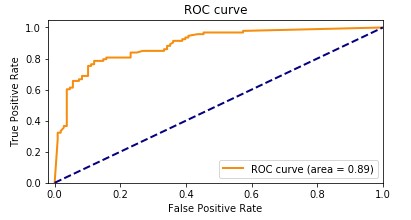
Our Amazon HealthLake augmented model had an accuracy of 89.1%. The following graph shows the ROC curve.
The following graph shows the confusion matrix.
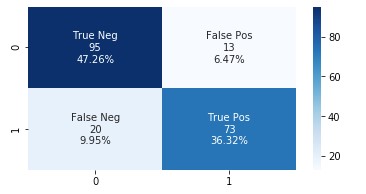
Adding the features extracted from Amazon HealthLake allowed us to improve the model accuracy from 85% to 89% and also the AUC from 0.86 to 0.89. If you look at the confusion matrices for the two models, the false positives reduced from 20 to 13 and the false negatives reduced from 27 to 20.
Optimizing healthcare is about ensuring the patient is associated with their peers and the right cohort. As patient data is added or changes, it’s important to continuously identify and reduce false negative and positive identifiers for overall improvement in the quality of care.
To better explain the performance improvements, we picked a patient from the false negative cohort in the first model who moved to true positive in the second model. We plotted a word cloud for the top medical conditions for this patient for the first and the second model, as shown in the following images.
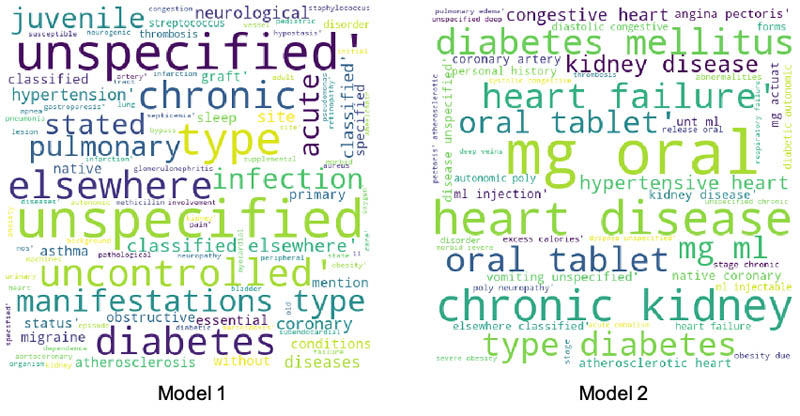
There is a clear difference between the medical conditions of the patient before and after the addition of features from Amazon HealthLake. The word cloud for model 2 is richer, with more medical conditions indicative of CHF than the one for model 1. The data embedded within the unstructured notes for this patient extracted by Amazon HealthLake helped this patient move from a false negative category to a true positive.
These numbers are based on synthetic experimental data we used from a subset of MIMIC-III patients. In a real-world scenario with higher-volume of patients, these numbers may differ.
Example 2: Grouping patients diagnosed with sepsis
For the second experiment, we took 500 patients with a positive sepsis diagnosis. We grouped these patients on the basis of their structured clinical records using k-means clustering. To show that this is a repeatable pattern, we chose the same feature engineering techniques as described in experiment 1. We didn’t divide the data into training and testing datasets because we were implementing an unsupervised learning algorithm.
We first analyzed the optimal number of clusters of the grouping using the Elbow method and arrived at the curve shown in the following graph.
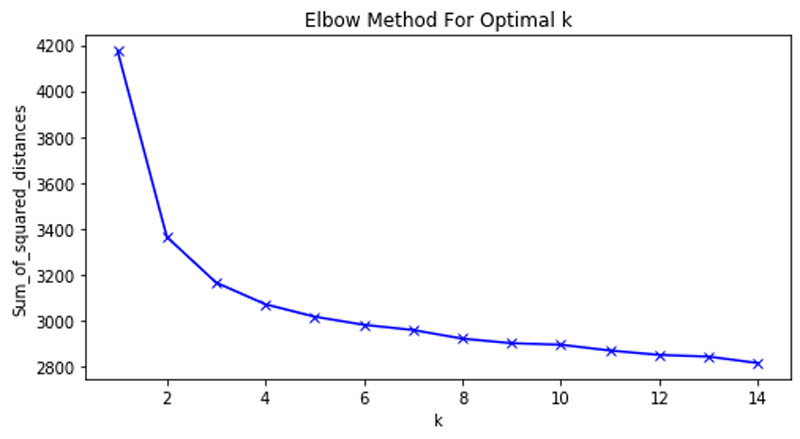
This allowed us to determine that six clusters were the optimal number in our patient grouping.
Baseline model
We reduced the dimensionality of the input data using Principal Component Analysis (PCA) to two and plotted the following scatter plot.
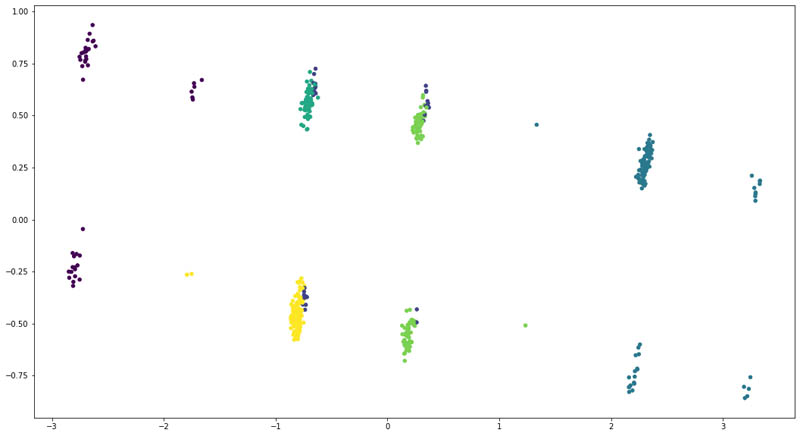
The following were the counts of patients across each cluster:
Cluster 1
Number of patients: 44
Cluster 2
Number of patients: 30
Cluster 3
Number of patients: 109
Cluster 4
Number of patients: 66
Cluster 5
Number of patients: 106
Cluster 6
Number of patients: 145
We found that the at least four of the six clusters had a distinct overlap of patients. That means the structured clinical features weren’t enough to clearly divide the patients into six groups.
Enhanced model
For the enhanced model, we added the ICD-10 codes and their corresponding descriptions for each patient as extracted from Amazon HealthLake. However, this time, we could see a clear separation of the patient groups.
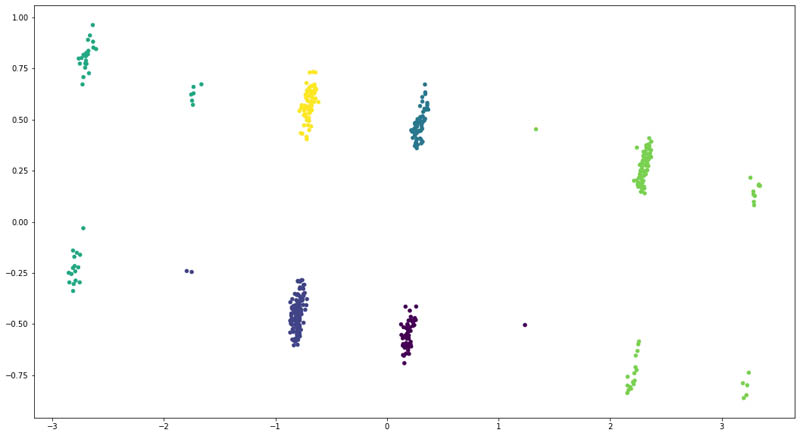
We also saw a change in distribution across the six clusters:
Cluster 1
Number of patients: 54
Cluster 2
Number of patients: 154
Cluster 3
Number of patients: 64
Cluster 4
Number of patients: 44
Cluster 5
Number of patients: 109
Cluster 6
Number of patients: 75
As you can see, adding features from the unstructured data for the patients allows us to improve the clustering model to clearly divide the patients into six clusters. We even saw that some patients moved across clusters, denoting that the model became better at recognizing those patients based on their unstructured clinical records.
Conclusion
In this post, we demonstrated how you can easily use SageMaker to build ML models on your data in Amazon HealthLake. We also demonstrated the advantages of augmenting data from unstructured clinical notes to improve the accuracy of disease prediction models. We hope this body of work provides you with examples of how to build ML models using SageMaker with your data stored and normalized in Amazon HealthLake and improve model performance for clinical outcome predictions. To learn more about Amazon HealthLake, please check the website and technical documentation for more information.
About the Authors
 Ujjwal Ratan is a Principal Machine Learning Specialist in the Global Healthcare and Lifesciences team at Amazon Web Services. He works on the application of machine learning and deep learning to real world industry problems like medical imaging, unstructured clinical text, genomics, precision medicine, clinical trials and quality of care improvement. He has expertise in scaling machine learning/deep learning algorithms on the AWS cloud for accelerated training and inference. In his free time, he enjoys listening to (and playing) music and taking unplanned road trips with his family.
Ujjwal Ratan is a Principal Machine Learning Specialist in the Global Healthcare and Lifesciences team at Amazon Web Services. He works on the application of machine learning and deep learning to real world industry problems like medical imaging, unstructured clinical text, genomics, precision medicine, clinical trials and quality of care improvement. He has expertise in scaling machine learning/deep learning algorithms on the AWS cloud for accelerated training and inference. In his free time, he enjoys listening to (and playing) music and taking unplanned road trips with his family.
 Nihir Chadderwala is an AI/ML Solutions Architect on the Global Healthcare and Life Sciences team. His background is building Big Data and AI-powered solutions to customer problems in variety of domains such as software, media, automotive, and healthcare. In his spare time, he enjoys playing tennis, watching and reading about Cosmos.
Nihir Chadderwala is an AI/ML Solutions Architect on the Global Healthcare and Life Sciences team. His background is building Big Data and AI-powered solutions to customer problems in variety of domains such as software, media, automotive, and healthcare. In his spare time, he enjoys playing tennis, watching and reading about Cosmos.
 Parminder Bhatia is a science leader in the AWS Health AI, currently building deep learning algorithms for clinical domain at scale. His expertise is in machine learning and large scale text analysis techniques in low resource settings, especially in biomedical, life sciences and healthcare technologies. He enjoys playing soccer, water sports and traveling with his family.
Parminder Bhatia is a science leader in the AWS Health AI, currently building deep learning algorithms for clinical domain at scale. His expertise is in machine learning and large scale text analysis techniques in low resource settings, especially in biomedical, life sciences and healthcare technologies. He enjoys playing soccer, water sports and traveling with his family.
