Discovering systematic errors with cross-modal embeddings
In this blog post, we introduce Domino, a new approach for discovering systematic errors made by machine learning models. We also discuss a framework for quantitatively evaluating methods like Domino.
Links:
📄 Paper (ICLR 2022)
🌍 Longer Walkthrough
💻 GitHub
📘 Docs
📒 Google Colab
Machine learning models that achieve high overall accuracy often make systematic errors on coherent slices of validation data.
What is a slice? A slice is a set of data samples that share a common characteristic. As an example, in large image datasets, photos of vintage cars comprise a slice (i.e. all images in the slice share a common subject). The term slice has a number of synonyms that you might be more familiar with (e.g. subgroup, subpopulation, stratum). These terms are largely interchangeable, but we’ll stick with “slice” throughout this post. We say that a model underperforms on a slice if performance on the data samples in the slice is significantly worse than its overall performance.
The search for underperforming slices is a critical, but often overlooked, part of model evaluation. When practitioners are aware of the slices on which their models underperform, they can make more informed decisions around model deployment. This is particularly important in safety-critical settings like medicine: a diagnostic model that underperforms on younger patients should likely not be deployed at a pediatric hospital. Slice awareness can also help practitioners debug and improve models: after an underperforming slice is identified, we can improve model robustness by either updating the training dataset or using robust optimization techniques (e.g. Sohoni et al., 2020; Sagawa et al., 2020).
Deploying models that underperform on critical data slices may have significant safety or fairness consequences. For example, models trained to detect collapsed lungs in chest X-rays have been shown to make predictions based on the presence of chest drains, a device typically used during treatment (Oakden-Rayner, 2019). As a result, these models often fail to detect collapsed lung in images without chest drains, a critical data slice where false negative predictions could be life-threatening.
However, in practice, some underperforming slices are hard to find. The examples in these “hidden” data slices are tied together by a concept not annotated in metadata or easily extracted from unstructured inputs (e.g. images, video, time-series data). Returning to our example from earlier, many chest X-ray datasets do not provide metadata indicating which patients’ images show chest tubes, making it difficult to evaluate performance on the slice. This raises the following question: How can we automatically identify data slices on which our model underperforms?
In this blog post, we discuss our recent exploration of this question. We introduce Domino, a novel method for identifying and describing underperforming slices. We also discuss an evaluation framework for rigorously evaluating our method across diverse slice types, tasks, and datasets.
What is slice discovery?
Slice discovery is the task of mining unstructured input data (e.g. images, videos, audio) for semantically meaningful subgroups on which a model performs poorly. We refer to automated techniques that mine input data for semantically meaningful slices as slice discovery methods (SDM). Given a labeled validation dataset and a trained classifier, an SDM computes a set of slicing functions that partition the dataset into slices. This process is illustrated below.
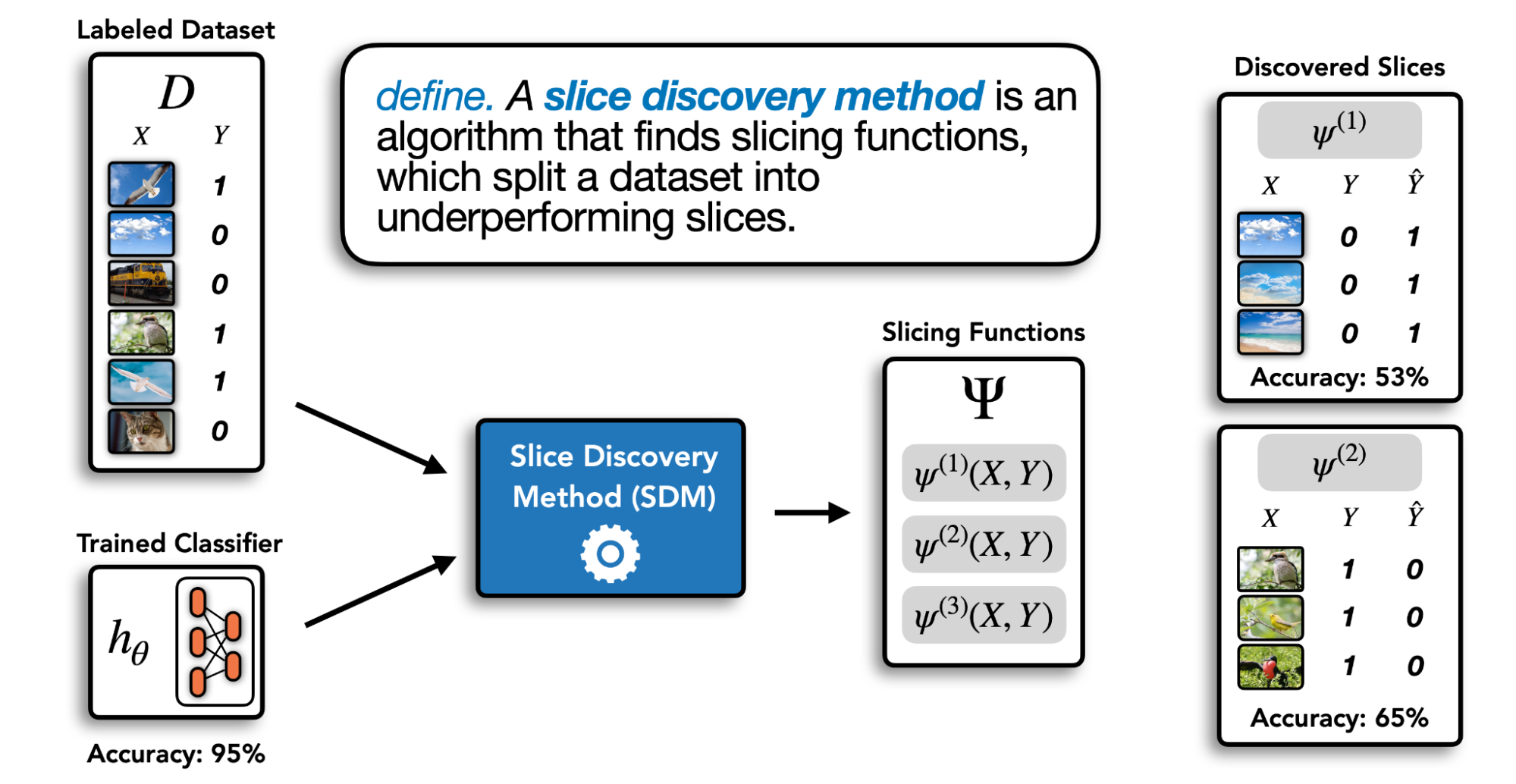
In order to be broadly useful across diverse settings, an ideal SDM should satisfy the following desiderata:
- Identified slices should contain examples on which the model underperforms, or has a high error rate.
- Identified slices should contain examples that are coherent, or align closely with a human-understandable concept.
This second desideratum is particularly hard to achieve: existing evaluations have shown that discovered slices often do not align with concepts understandable to a domain expert. Further, even if slices do align well with concepts, it may be difficult for humans to identify the commonality.
Domino: Slice discovery with cross-modal embeddings
In our work, we introduce Domino, a slice discovery method designed to identify coherent, underperforming data slices (i.e. groups of similar validation data points on which the model makes errors). It leverages a powerful class of recently-developed cross-modal representation learning approaches, which yield semantically-meaningful representations by embedding images and text in the same latent space. We demonstrate that using cross-modal representations both improves slice coherence and enables Domino to generate natural language descriptions for identified slices!
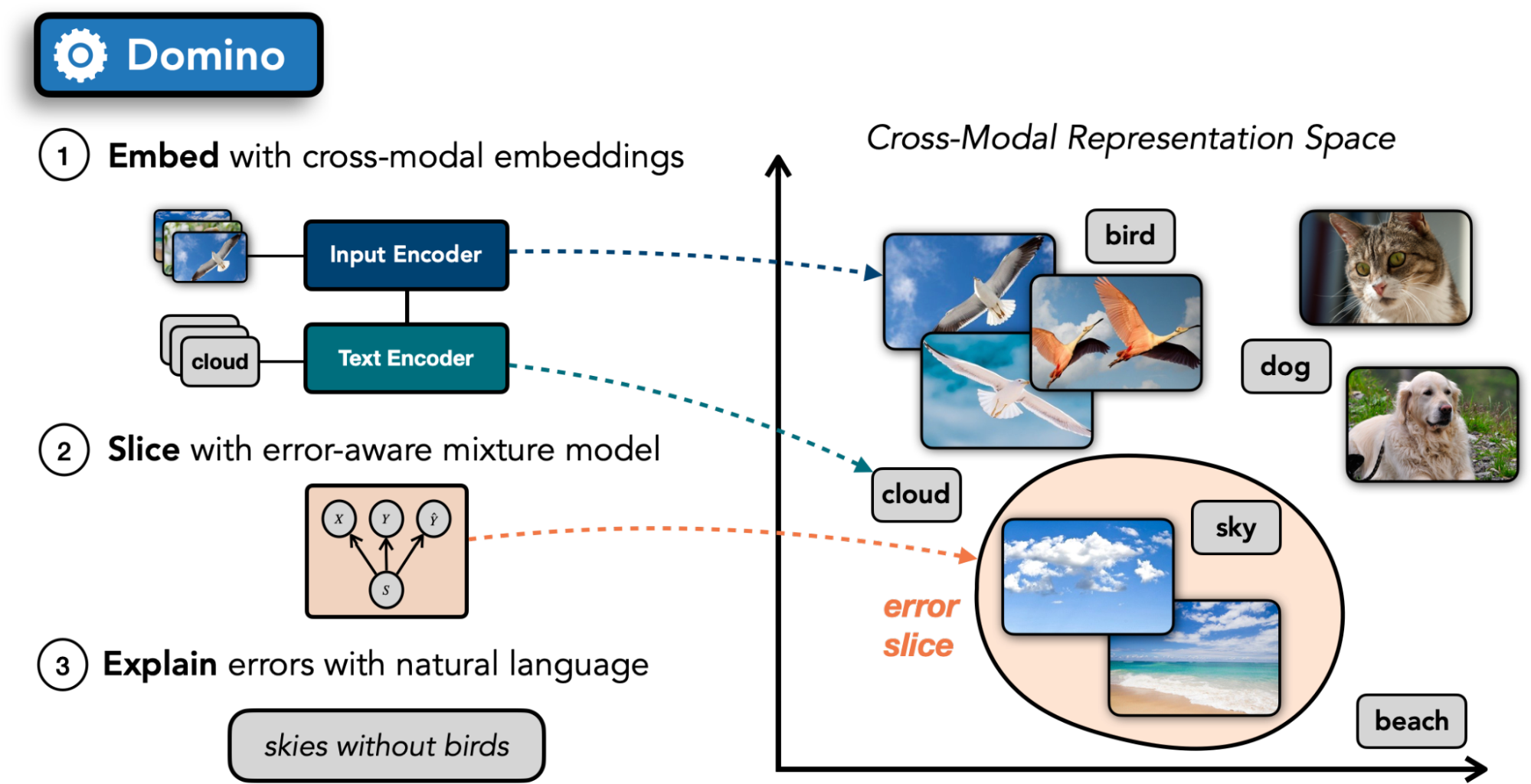
Domino follows a three-step procedure illustrated in the figure above:
- Embed: Domino encodes the validation images alongside text in a shared embedding space using a cross-modal encoder. In many domains, such encoders are publicly available (e.g. CLIP for natural images, VideoCLIP for natural videos, ConVIRT for medical images, and CLASP for amino acid sequences).
- Slice: Using an error-aware mixture model, Domino identifies regions in the embedding space with a high concentration of errors.
- Describe: Finally, to help practitioners understand the commonalities among the examples in each slice, Domino generates natural language descriptions of the slices. To do so, it leverages the cross-modal embeddings computed in Step 1, surfacing the text nearest to the slice in embedding space.
We now use Domino to audit a popular off-the-shelf classifier: a ResNet18 pretrained on ImageNet. Specifically, we interrogate the model’s ability to detect cars, exploring whether there are any interesting slices on which the model underperforms. In the figure below we show a couple of the slices that Domino discovered. The gray boxes show the natural language descriptions of the two slices produced by Domino, the $X$ row shows the top six images predicted by domino to be in the slice, the $Y$ row shows the ground truth label assigned to the image, and the $hat{Y}$ row shows the ResNet18’s predicted probability for the “car” class. Note that although we only include six images here, the complete slice includes dozens of images.

From these slices, we might hypothesize that the model struggles to recognize photos of cars taken from the inside and photos of racecars. Both of these slices describe rare subclasses of the target class. Depending on the intended use case for the model, we may want to add more training examples to boost performance in these slices. For example, Waymo (an autonomous vehicle company) may not care much whether the model misses photos of car interiors, but ESPN (a broadcaster with the television rights for Formula 1) would care a lot if the model couldn’t recognize race cars! Clearly, it’s important to practitioners that discovered slices map onto coherent concepts.
Evaluating slice discovery methods
In designing Domino, we were inspired by a number of really exciting slice discovery methods that were recently proposed. These include The Spotlight (D’Eon et al. 2022), GEORGE (Sohoni et al. 2020), and MultiAccuracy Boost (Kim et al. 2018). These methods all have (1) an embed step and (2) a slice step, like Domino, but use different embeddings and slicing algorithms. In our experiments, we evaluate SDMs along these two axes, ablating both the choice of embedding and the slicing algorithm. Notably, these methods do not include a (3) describe step, and generally require users to manually inspect examples and identify common attributes.
SDMs like Domino have traditionally been evaluated qualitatively, due to a lack of a simple quantitative approach. Typically, in these evaluations, the SDM is applied to a few different models and identified slices are visualized. Practitioners can then inspect the slices and judge whether the slices “make sense.” However, these qualitative evaluations are subjective and do not scale beyond more than a few settings. Moreover, they cannot tell us if the SDM has missed an important, coherent slice.
Ideally, we’d like to estimate the failure rate of an SDM: how often it fails to identify a coherent slice on which the model underperforms. Estimating this failure rate is very challenging because we don’t typically know the full set of slices on which a model underperforms. How could we possibly know if the SDM is missing any?
To solve this problem, we trained 1,235 deep classifiers that were specifically constrained to underperform on pre-defined slices. We did this across three domains: natural images, medical images and medical time-series. Our approach involved (1) obtaining a dataset with some annotated slices (e.g. a dataset with interesting annotated attributes, like CelebA or MIMIC-CXR), and (2) manipulating the dataset such that, with high probability, a model trained on it would exhibit poor performance on one or more of the annotated slices (e.g. by subsampling the dataset to induce a spurious correlation between the label and a metadata field).
Using this evaluation framework, we were able to evaluate Domino quantitatively. We find that Domino accurately identifies 36% of the 1,235 slices in our framework. Further, the natural language description of the generated slice exactly matches the name of the slice in 35% of settings.
We were also able to compare SDMs and run ablation studies evaluating specific SDM design choices. Two key findings emerged from these experiments:
- Cross-modal embeddings improve SDM performance. We found that the choice of representation matters – a lot! Slice discovery methods based on cross-modal embeddings outperform those based on a single modality by at least 9 percentage-points in precision-at-10. When compared with using the activations of the trained model, the gap grows to 15 percentage points. This finding is of particular interest given that classifier activations are a popular embedding choice in existing SDMs.
- Modeling both the prediction and class label enables accurate slice discovery. Good embeddings alone do not suffice – the choice of algorithm for actually extracting the underperforming slices from the embedding space, is significant as well. We find that a simple mixture model that jointly models the embeddings, labels and predictions enables a 105% improvement over the next best slicing algorithm. We hypothesize that this is because this algorithm is unique in modeling the class labels and the model’s predictions as separate variables, which leads to slices which are “pure” in their error type (false positive vs. false negative).
However, there’s still a long way to go: slice discovery is a challenging task, and Domino, the best performing method in our experiments, still fails to recover over 60% of coherent slices. We see a number of exciting avenues for future work that could begin to close this gap.
-
We suspect that improvements in the embeddings that power slice discovery will be driven by large cross modal datasets, so work in dataset curation and management could help push the needle.
-
In this blog post, we described slice discovery as a fully automated process, while, in the future, we expect that effective slice discovery systems will be highly interactive: practitioners will be able to quickly explore slices and provide feedback. Forager, a system for rapid data exploration, is an exciting step in this direction.
We are really excited to continue working on this important problem and to collaborate with others as we seek to develop more reliable slice discovery methods. To facilitate this process, we are releasing 984 models and their associated slices as part of dcbench, a suite of data centric benchmarks. This will allow others to reproduce our results and also develop new slice discovery methods. Additionally, we are also releasing domino, a Python package containing implementations of popular slice discovery methods. If you’ve developed a new slice discovery method and would like us to add it to the library please reach out!
