The process of building a machine learning (ML) model is iterative until you find the candidate model that is performing well and is ready to be deployed. As data scientists iterate through that process, they need a reliable method to easily track experiments to understand how each model version was built and how it performed.
Amazon SageMaker allows teams to take advantage of a broad range of features to quickly prepare, build, train, deploy, and monitor ML models. Amazon SageMaker Pipelines provides a repeatable process for iterating through model build activities, and is integrated with Amazon SageMaker Experiments. By default, every SageMaker pipeline is associated with an experiment, and every run of that pipeline is tracked as a trial in that experiment. Then your iterations are automatically tracked without any additional steps.
In this post, we take a closer look at the motivation behind having an automated process to track experiments with Experiments and the native capabilities built into Pipelines.
Why is it important to keep your experiments organized?
Let’s take a step back for a moment and try to understand why it’s important to have experiments organized for machine learning. When data scientists approach a new ML problem, they have to answer many different questions, from data availability to how they will measure model performance.
At the start, the process is full of uncertainty and is highly iterative. As a result, this experimentation phase can produce multiple models, each created from their own inputs (datasets, training scripts, and hyperparameters) and producing their own outputs (model artifacts and evaluation metrics). The challenge then is to keep track of all these inputs and outputs of each iteration.
Data scientists typically train many different model versions until they find the combination of data transformation, algorithm, and hyperparameters that results in the best performing version of a model. Each of these unique combinations is a single experiment. With a traceable record of the inputs, algorithms, and hyperparameters that were used by that trial, the data science team can find it easy to reproduce their steps.
Having an automated process in place to track experiments improves the ability to reproduce as well as deploy specific model versions that are performing well. The Pipelines native integration with Experiments makes it easy to automatically track and manage experiments across pipeline runs.
Benefits of SageMaker Experiments
SageMaker Experiments allows data scientists organize, track, compare, and evaluate their training iterations.
Let’s start first with an overview of what you can do with Experiments:
- Organize experiments – Experiments structures experimentation with a top-level entity called an experiment that contains a set of trials. Each trial contains a set of steps called trial components. Each trial component is a combination of datasets, algorithms, and parameters. You can picture experiments as the top-level folder for organizing your hypotheses, your trials as the subfolders for each group test run, and your trial components as your files for each instance of a test run.
- Track experiments – Experiments allows data scientists to track experiments. It offers the possibility to automatically assign SageMaker jobs to a trial via simple configurations and via the tracking SDKs.
- Compare and evaluate experiments – The integration of Experiments with Amazon SageMaker Studio makes it easy to produce data visualizations and compare different trials. You can also access the trial data via the Python SDK to generate your own visualization using your preferred plotting libraries.
To learn more about Experiments APIs and SDKs, we recommend the following documentation: CreateExperiment and Amazon SageMaker Experiments Python SDK.
If you want to dive deeper, we recommend looking into the amazon-sagemaker-examples/sagemaker-experiments GitHub repository for further examples.
Integration between Pipelines and Experiments
The model building pipelines that are part of Pipelines are purpose-built for ML and allow you to orchestrate your model build tasks using a pipeline tool that includes native integrations with other SageMaker features as well as the flexibility to extend your pipeline with steps run outside SageMaker. Each step defines an action that the pipeline takes. The dependencies between steps are defined by a direct acyclic graph (DAG) built using the Pipelines Python SDK. You can build a SageMaker pipeline programmatically via the same SDK. After a pipeline is deployed, you can optionally visualize its workflow within Studio.
Pipelines automatically integrate with Experiments by automatically creating an experiment and trial for every run. Pipelines automatically create an experiment and a trial for every run of the pipeline before running the steps unless one or both of these inputs are specified. While running the pipeline’s SageMaker job, the pipeline associates the trial with the experiment, and associates to the trial every trial component that is created by the job. Specifying your own experiment or trial programmatically allows you to fine-tune how to organize your experiments.
The workflow we present in this example consists of a series of steps: a preprocessing step to split our input dataset into train, test, and validation datasets; a tuning step to tune our hyperparameters and kick off training jobs to train a model using the XGBoost built-in algorithm; and finally a model step to create a SageMaker model from the best trained model artifact. Pipelines also offers several natively supported step types outside of what is discussed in this post. We also illustrate how you can track your pipeline workflow and generate metrics and comparison charts. Furthermore, we show how to associate the new trial generated to an existing experiment that might have been created before the pipeline was defined.
SageMaker Pipelines code
You can review and download the notebook from the GitHub repository associated with this post. We look at the Pipelines-specific code to understand it better.
Pipelines enables you to pass parameters at run time. Here we define the processing and training instance types and counts at run time with preset defaults:
Next, we set up a processing script that downloads and splits the input dataset into train, test, and validation parts. We use SKLearnProcessor for running this preprocessing step. To do so, we define a processor object with the instance type and count needed to run the processing job.
Pipelines allows us to achieve data versioning in a programmatic way by using execution-specific variables like ExecutionVariables.PIPELINE_EXECUTION_ID, which is the unique ID of a pipeline run. We can, for example, create a unique key for storing the output datasets in Amazon Simple Storage Service (Amazon S3) that ties them to a specific pipeline run. For the full list of variables, refer to Execution Variables.
Then we move on to create an estimator object to train an XGBoost model. We set some static hyperparameters that are commonly used with XGBoost:
We do hyperparameter tuning of the models we create by using a ContinuousParameter range for lambda. Choosing one metric to be the objective metric tells the tuner (the instance that runs the hyperparameters tuning jobs) that you will evaluate the training job based on this specific metric. The tuner returns the best combination based on the best value for this objective metric, meaning the best combination that minimizes the best root mean square error (RMSE).
The tuning step runs multiple trials with the goal of determining the best model among the parameter ranges tested. With the method get_top_model_s3_uri, we rank the top 50 performing versions of the model artifact S3 URI and only extract the best performing version (we specify k=0 for the best) to create a SageMaker model.
When the pipeline runs, it creates trial components for each hyperparameter tuning job and each SageMaker job created by the pipeline steps.
You can further configure the integration of pipelines with Experiments by creating a PipelineExperimentConfig object and pass it to the pipeline object. The two parameters define the name of the experiment that will be created, and the trial that will refer to the whole run of the pipeline.
If you want to associate a pipeline run to an existing experiment, you can pass its name, and Pipelines will associate the new trial to it. You can prevent the creation of an experiment and trial for a pipeline run by setting pipeline_experiment_config to None.
We pass on the instance types and counts as parameters, and chain the preceding steps in order as follows. The pipeline workflow is implicitly defined by the outputs of a step being the inputs of another step.
The full-fledged pipeline is now created and ready to go. We add an execution role to the pipeline and start it. From here, we can go to the SageMaker Studio Pipelines console and visually track every step. You can also access the linked logs from the console to debug a pipeline.
The preceding screenshot shows in green a successfully run pipeline. We obtain the metrics of one trial from a run of the pipeline with the following code:
Compare the metrics for each trial component
You can plot the results of hyperparameter tuning in Studio or via other Python plotting libraries. We show both ways of doing this.
Explore the training and evaluation metrics in Studio
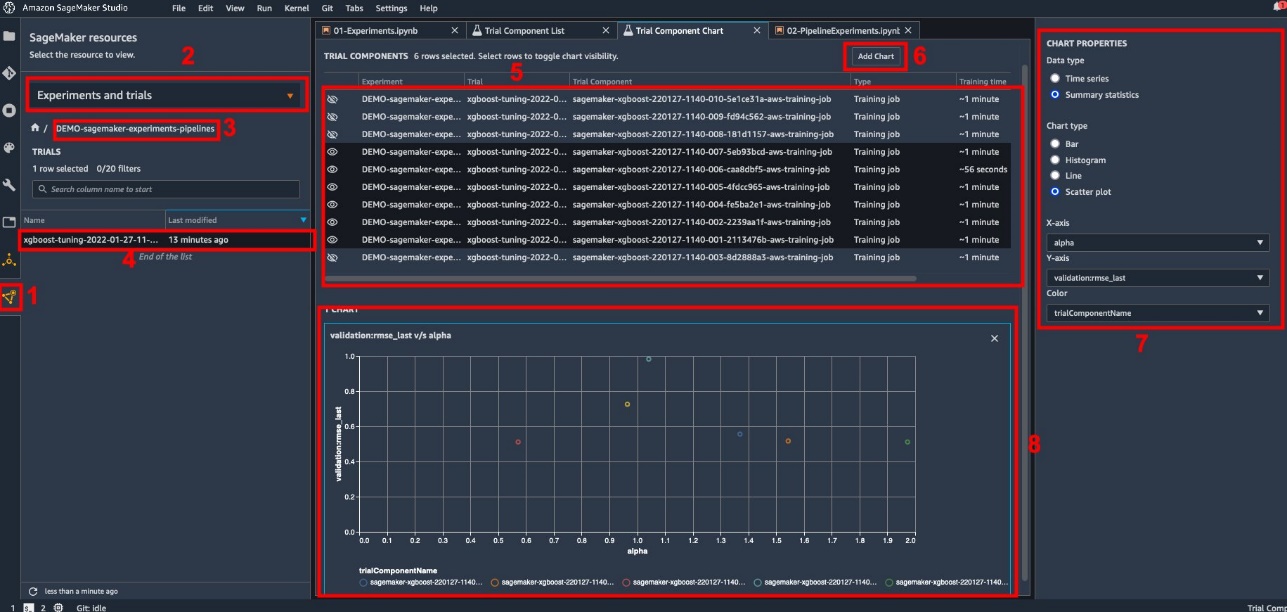
Studio provides an interactive user interface where you can generate interactive plots. The steps are as follows:
- Choose Experiments and Trials from the SageMaker resources icon on the left sidebar.
- Choose your experiment to open it.
- Choose (right-click) the trial of interest.
- Choose Open in trial component list.
- Press Shift to select the trial components representing the training jobs.
- Choose Add chart.
- Choose New chart and customize it to plot the collected metrics that you want to analyze. For our use case, choose the following:
- For Data type¸ select Summary Statistics.
- For Chart type¸ select Scatter Plot.
- For X-axis, choose
lambda. - For Y-axis, choose
validation:rmse_last.
The new chart appears at the bottom of the window, labeled as ‘8’.
You can include more or fewer training jobs by pressing Shift and choosing the eye icon for a more interactive experience.
Analytics with SageMaker Experiments
When the pipeline run is complete, we can quickly visualize how different variations of the model compare in terms of the metrics collected during training. Earlier, we exported all trial metrics to a Pandas DataFrame using ExperimentAnalytics. We can reproduce the plot obtained in Studio by using the Matplotlib library.
Conclusion
The native integration between SageMaker Pipelines and SageMaker Experiments allows data scientists to automatically organize, track, and visualize experiments during model development activities. You can create experiments to organize all your model development work, such as the following:
- A business use case you’re addressing, such as creating an experiment to predict customer churn
- An experiment owned by the data science team regarding marketing analytics, for example
- A specific data science and ML project
In this post, we dove into Pipelines to show how you can use it in tandem with Experiments to organize a fully automated end-to-end workflow.
As a next step, you can use these three SageMaker features – Studio, Experiments and Pipelines – for your next ML project.
Suggested readings
- Amazon SageMaker now supports cross-account lineage tracking and multi-hop lineage querying
- Announcing Amazon SageMaker Inference Recommender
- Introducing the Well-Architected Framework for Machine Learning
- Machine Learning Lens: AWS Well-Architected Framework
- Roundup of re:Invent 2021 Amazon SageMaker announcements
About the authors
 Paolo Di Francesco is a solutions architect at AWS. He has experience in the telecommunications and software engineering. He is passionate about machine learning and is currently focusing on using his experience to help customers reach their goals on AWS, in particular in discussions around MLOps. Outside of work, he enjoys playing football and reading.
Paolo Di Francesco is a solutions architect at AWS. He has experience in the telecommunications and software engineering. He is passionate about machine learning and is currently focusing on using his experience to help customers reach their goals on AWS, in particular in discussions around MLOps. Outside of work, he enjoys playing football and reading.
 Mario Bourgoin is a Senior Partner Solutions Architect for AWS, an AI/ML specialist, and the global tech lead for MLOps. He works with enterprise customers and partners deploying AI solutions in the cloud. He has more than 30 years of experience doing machine learning and AI at startups and in enterprises, starting with creating one of the first commercial machine learning systems for big data. Mario spends his free time playing with his three Belgian Tervurens, cooking dinner for his family, and learning about mathematics and cosmology.
Mario Bourgoin is a Senior Partner Solutions Architect for AWS, an AI/ML specialist, and the global tech lead for MLOps. He works with enterprise customers and partners deploying AI solutions in the cloud. He has more than 30 years of experience doing machine learning and AI at startups and in enterprises, starting with creating one of the first commercial machine learning systems for big data. Mario spends his free time playing with his three Belgian Tervurens, cooking dinner for his family, and learning about mathematics and cosmology.
 Ganapathi Krishnamoorthi is a Senior ML Solutions Architect at AWS. Ganapathi provides prescriptive guidance to startup and enterprise customers helping them to design and deploy cloud applications at scale. He is specialized in machine learning and is focused on helping customers leverage AI/ML for their business outcomes. When not at work, he enjoys exploring outdoors and listening to music.
Ganapathi Krishnamoorthi is a Senior ML Solutions Architect at AWS. Ganapathi provides prescriptive guidance to startup and enterprise customers helping them to design and deploy cloud applications at scale. He is specialized in machine learning and is focused on helping customers leverage AI/ML for their business outcomes. When not at work, he enjoys exploring outdoors and listening to music.
 Valerie Sounthakith is a Solutions Architect for AWS, working in the Gaming Industry and with Partners deploying AI solutions. She is aiming to build her career around Computer Vision. During her free time, Valerie spends it to travel, discover new food spots and change her house interiors.
Valerie Sounthakith is a Solutions Architect for AWS, working in the Gaming Industry and with Partners deploying AI solutions. She is aiming to build her career around Computer Vision. During her free time, Valerie spends it to travel, discover new food spots and change her house interiors.