Daphne Koller is the CEO and founder of Insitro, a machine learning-driven drug discovery and development company that recently made news for its identification of a novel drug target for ALS and its collaboration with Eli Lilly to license Lilly’s biochemical delivery systems. Prior to founding Insitro, Daphne was the co-founder, co-CEO, and president of the online education platform Coursera.
Noubar Afeyan is the founder and CEO of Flagship Pioneering, which creates biotechnology companies focused on transforming human health and environmental sustainability. He is also co-founder and chairman of the messenger RNA company Moderna. An entrepreneur and biochemical engineer, Noubar has numerous patents to his name and has co-founded many startups in science and technology.
Dr. Eric Topol is the executive vice president of the biomedical research non-profit Scripps Research, where he founded and now directs the Scripps Research Translational Institute. One of the most cited researchers in medicine, Eric has focused on promoting human health and individualized medicine through the use of genomic and digital data and AI.
These three are likely to have an outsized influence on how drugs and new medical technologies soon will be developed.
[TRANSITION MUSIC]
Here’s my interview with Daphne Koller:
LEE: Daphne, I’m just thrilled to have you join us.
DAPHNE KOLLER: Thank you for having me, Peter. It’s a pleasure to be here.
LEE: Well, you know, you’re quite well-known across several fields. But maybe for some audience members of this podcast, they might not have encountered you before. So where I’d like to start is a question I’ve been asking all of our guests.
How would you describe what you do? And the way I kind of put it is, you know, how do you explain to someone like your parents what you do for a living?
KOLLER: So that answer obviously has shifted over the years.
What I would say now is that we are working to leverage the incredible convergence of very powerful technologies, of which AI is one but not the only one, to change the way in which we discover and develop new treatments for diseases for which patients are currently suffering and even dying.
LEE: You know, I think I’ve known you for a long time.
KOLLER: Longer than I think either of us care to admit.
LEE: [LAUGHS] In fact, I think I remember you even when you were still a graduate student. But of course, I knew you best when you took up your professorship at Stanford. And I always, in my mind, think of you as a computer scientist and a machine learning person. And in fact, you really made a big name for yourself in computer science research in machine learning.
But now you’re, you know, leading one of the most important biotech companies on the planet. How did that happen?
KOLLER: So people often think that this is a recent transition. That is, after I left Coursera, I looked around and said, “Hmm. What should I do next? Oh, biotech seems like a good thing,” but that’s actually not the way it transpired.
This goes all the way back to my early days at Stanford, where, in fact, I was, you know, as a young faculty member in machine learning, because I was the first machine learning hire into Stanford’s computer science department, I was looking for really exciting places in which this technology could be deployed, and applications back then, because of scarcity of data, were just not that inspiring.
And so I looked around, and this was around the late ’90s, and realized that there was interesting data emerging in biology and medicine. My first application actually was in, interestingly, in epidemiology—patient tracking and tuberculosis. You know, you can think of it as a tiny microcosm of the very sophisticated models that COVID then enabled in a much later stage.
LEE: Right.
KOLLER: And so initially, this was based almost entirely on just technical interest. It’s kind of like, oh, this is more interesting as a question to tackle than spam filtering. But then I became interested in biology in its own right, biology and medicine, and ended up having a bifurcated existence as a Stanford professor where half my lab continued to do core computer science research published in, you know, NeurIPS and ICML. And the other half actually did biomedical research that was published in, you know, Nature Cell [and] Science. So that was back in, you know, the early, early 2000s, and for most of my Stanford career, I continued to have both interests.
And then the Coursera experience kind of took me out of Stanford and put me in an industry setting for the first time in my life actually. But then when my time at Coursera came to an end, you know, I’d been there for five years. And if you look at the timeline, I left Stanford in early 2012, right as the machine learning revolution was starting. So I missed the beginning.
And it was only in like 2016 or so that, as I picked my head up over the trenches, like, “Oh my goodness, this technology is going to change the world.” And I wanted to deploy that big thing towards places where it would have beneficial impact on the world, like to make the world a better place.
LEE: Yeah.
KOLLER: And so I decided that one of the areas where I could make a unique, differentiated impact was in really bringing AI and machine learning to the life sciences, having spent, you know, the majority of my career at the boundary of those two disciplines. And notice I say “boundary” with deliberation because there wasn’t very much of an intersection.
LEE: Right.
KOLLER: I felt like I could do something that was unique.
LEE: So just to stick on you for a little bit longer, you know, we have been sort of getting into your origin story about what we call AI today—but machine learning, so deep learning.
And, you know, there has always been a kind of an emotional response for people like you and me and now the general public about their first encounters with what we now call generative AI. I’d love to hear what your first encounter was with generative AI and how you reacted to this.
KOLLER: I think my first encounter was actually an indirect one. Because, you know, the earlier generations of generative AI didn’t directly touch our work at Insitro (opens in new tab).
And yet at the same time, I had always had an interest in computer vision. That was a large part of my non-bio work when I was at Stanford.
And so some of my earlier even presentations, when I was trying to convey to people back in 2016 how this technology was going to transform the world, I was talking about the incredible progress in image recognition that had happened up until that point.
So my first interaction was actually in the generative AI for images, where you are able to go the other way …
LEE: Yes.
KOLLER: … where you can take a verbal description of an image and create—and this was back in the days when the images weren’t particularly photorealistic, but still a natural language description to an image was magic given that only two or three years before that, we were barely able to look at an image and write a short phrase saying, “This is a dog on the beach.” And so that arc, that hockey curve, was just mind blowing to me.
LEE: Did you have moments of skepticism?
KOLLER: Yeah, I mean the early, you know, early versions of ChatGPT, where it was more like parlor tricks and poking it a little bit revealed all of the easy ways that one could break it and make it do really stupid things. I was like, yeah, OK, this is kind of cute, but is it going to actually make a difference? Is it going to solve a problem that matters?
And I mean, obviously, I think now everyone agrees that the answer is yes, although there are still people who are like, yeah, but maybe it’s around the edges. I’m not among them, by the way, but … yeah, so initially there were like, “Yeah, this is cute and very impressive, but is it going to make a difference to a problem that matters?”
LEE: Yeah. So now, maybe this is a good time to get into what you’ve been doing with ALS [amyotrophic lateral sclerosis]. You know, there’s a knee-jerk reaction from the technology side to focus on designing small molecules, on predicting, you know, their properties, you know, maybe binding affinity or aspects of ADME [absorption, distribution, metabolism, and excretion], you know, like absorption or dispersion or whatever.
And all of that is very useful, but if I understand the work on ALS, you went to a much harder place, which is to actually identify and select targets.
KOLLER: That’s right.
LEE: So first off, just for the benefit of the standard listeners of this podcast, explain what that problem is in general.
KOLLER: No, for sure. And I think maybe I’ll start by just very quickly talking about the drug discovery and development arc, …
LEE: Yeah.
KOLLER: … which, by and large, consists of three main phases. That’s the standard taxonomy. The first is what’s called sometimes target discovery or identifying a therapeutic hypothesis, which looks like: if I modulate this target in this disease, something beneficial will happen.
Then, you have to take that target and turn it into a molecule that you can actually put into a person. It could be a small molecule. It could be a large molecule like an antibody, whatever. And then you have that construct, that molecule. And the last piece is you put it into a person in the context of a clinical trial, and you measure what has happened. And there’s been AI deployed towards each of those three stages in different ways.
The last one is mostly like an efficiency gain. You know, the trial is kind of already defined, and you want to deploy technology to make it more efficient and effective, which is great because those are expensive operations.
LEE: Yep.
KOLLER: The middle one is where I would say the vast majority of efforts so far has been deployed in AI because it is a nice, well-defined problem. It doesn’t mean it’s easy, but it’s one where you can define the problem. It is, I need to inhibit this protein by this amount, and the molecule needs to be soluble and whatever and go past the blood-brain barrier. And you know probably within a year and a half or so, or two, if you succeeded or not.
The first stage is the one where I would say the least amount of energy has gone because when you’re uncovering a novel target in the context of an indication, you don’t know that you’ve been successful until you go all the way to the end, which is the clinical trial, which is what makes this a long and risky journey. And not a lot of people have the appetite or the capital to actually do that.
However, in my opinion, and that of, I think, quite a number of others, it is where the biggest impact can be made. And the reason is that while pharma has its deficiencies, making good molecules is actually something they’re pretty good at.
It might take them longer than it should, maybe it’s not as efficient as it could be, but at the end of the day, if you tell them to drug A target, pharma is actually pretty good at generating those molecules. However, when you put those molecules into the clinic, 90% of them fail. And the reason they fail is not by and large because the molecule wasn’t good. In the majority of cases, it’s because the target you went after didn’t do anything useful in the context of the patient population in which you put it.
And so in order to fix the inefficiency of this industry, which is incredible inefficiency, you need to address the problem at the root, and the root is picking the right targets to go after. And so that is what we elected to do.
It doesn’t mean we don’t make molecules. I mean, of course, you can’t just end up with a target because a target is not actionable. You need to turn it into a molecule. And we absolutely do that. And by the way, the partnership with Lilly (opens in new tab) is actually one where they help us make a molecule.
LEE: Yes.
KOLLER: I mean, it’s our target. It’s our program. But Lilly is deploying its very state-of-the-art molecule-making capabilities to help us turn that target into a drug.
LEE: So let’s get now into the machine learning of this. Again, this just strikes me as such a difficult problem to solve.
KOLLER: Yeah.
LEE: So how does machine learning … how does AI help you?
KOLLER: So I think when you look at how people currently select targets, it’s a combination of oftentimes at this point, with an increasing respect for the power of human genetics, some search for a genetic association, oftentimes with a human-defined, highly subjective, highly noisy clinical outcome, like some ICD [International Classification of Diseases] code.
And those are often underpowered and very difficult to deconvolute the underlying biology. You combine that with some mechanistic interrogation in a highly reductionist model system looking at a small number of readouts, biochemical readouts, that a biologist thinks are relevant to the disease. Like does this make this, whatever, cholesterol go up or amyloid beta go down? Or whatever. And then you take that as the second stage, and you pick, based on typically human intuition about, Oh, this one looks good to me, and then you take that forward.
What we’re doing is an attempt to be as unbiased and holistic as possible. So, first of all, rather than rely on human-defined clinical endpoints, like this person has been diagnosed with diabetes or fatty liver, we try and measure as much as we can a holistic physiological state and then use machine learning to find structure, patterns in that human physiological readouts, imaging readouts, and omics readouts from blood, from tissue, different kinds of imaging, and say, these are different vectors that this disease takes, this group of individuals, and here’s a different group of individuals that maybe from a diagnostical perspective are all called the same thing, but they are actually exhibiting a very different biology underlying it.
And so that is something that doesn’t emerge when a human being takes a reductionist view to looking at this high-content data, and oftentimes, they don’t even look at it and produce an ICD code.
LEE: Right. Yep.
KOLLER: The same approach, actually even the same code base, is taken in the cellular data. So we don’t just say, “Well, the thing that matters is, you know, the total amount of lipid in the cell or whatever.” Rather, we say, “Let’s look at multiple readouts, multiple ways of looking at the cells, combine them using the power of machine learning.” And again, looking at imaging readouts where a human’s eyes just glaze over looking at even a few dozen cells, far less a few hundreds of millions of cells, and understand what are the different biological processes that are going on. What are the vectors that the disease might take you in this direction, in this group of cells, or in that direction?
And then importantly, we take all of that information from the human side, from the cellular side, across these different readouts, and we combine them using an integrative approach that looks at the combined weight of evidence and says, these are the targets that I have the greatest amount of conviction about by looking across all of that information. Whereas we know, and we know this, I’m sure you’ve seen this analysis done for clinicians, a human being typically is able to keep three or four things in their head at the same time.
LEE: Right.
KOLLER: A really good human being who’s really expert at what they do can maybe get to six to eight.
LEE: Yeah.
KOLLER: The machine learning has no problem doing a few hundred.
LEE: Right.
KOLLER: And so you put that together, and that allows you, to your earlier question, really select the targets around which you have the highest conviction. And then those are the ones that we then prioritize for interrogation in more expensive systems like mice and monkeys and then at the end of the day pick the small handful that one can afford to actually take into clinical trials.
LEE: So now, Insitro recently received $25 million in milestone payments from Bristol Myers Squibb (opens in new tab) after discovering and selecting a novel drug target for ALS. Can you tell us a little bit more about that?
KOLLER: We are incredibly excited about the first novel target, and there is a couple of others just behind it in line that seem, you know, quite efficacious, as well, that truly seem to reverse, albeit in a cellular system, what we now understand to be ALS pathology across multiple different dimensions. There’s been obviously many attempts made to try and address ALS, which by the way, horrible, horrible disease, worse than most cancers. It kills you almost inevitably in three to five years in a particularly horrific way.
And what we have in our hands is a target that seems to revert a lot of the pathologies that are associated with the disease, which we now understand has to do with the mis-splicing of multiple proteins within the cell and creating defective versions of those proteins that are just not operational. And we are seeing reversion of many of those.
So can I tell you for sure it’ll work in a human? No, there’s many steps between now and then. But we couldn’t be more excited about the opportunity to provide what we hope will be a disease-modifying intervention for these patients who really desperately need something.
LEE: Well, it’s certainly been making waves in the biotech and biomedical world.
KOLLER: Thank you.
LEE: So we’ll be really watching very closely.
So, you know, I think just reflecting on, you know, what we missed and what we got right in our book, I think in our book, we did have the insight that there would be an ability to connect, say, genotypic and phenotypic data and, you know, just broadly the kinds of clinical measurements that get made on real patients and that these things could be brought together. And I think the work that you’re doing really illustrates that in a very, very sophisticated, very ambitious way.
But the fact that this could be connected all the way down to the biology, to the biochemistry, I think we didn’t have any clue what would happen, at least not this quickly.
KOLLER: Well, I think the …
LEE: And I realize, you’ve been at this for quite a few years, but still, it’s quite amazing.
KOLLER: The thread that connects them is human genetics. And I think that has, to us, been, sort of, the, kind of, the connective tissue that allows you to translate across different systems and say, “What does this gene do? What does this gene do in this organ and in that organ? What does it do in this type of cell and in that type of cell?”
And then use that as sort of the thread, if you will, that follows the impact of modulating this gene all the way from the simple systems where you can do the experiment to the complex systems where you can’t do the experiment until the very end, but you have the human genetics as a way of looking at the statistics and understanding what the impact might be.
LEE: So I’d like to now switch gears and take … I want to take two steps in the remainder of this conversation towards the future. So one step into that future, of course, we’re living through now, which is just all of the crazy pace of work and advancement in generative AI generally, you know, just the scale of transformers, of post-training, and now inference scale and reasoning models and so on. And where do you see all of that going with respect to the goals that you have and that Insitro has?
KOLLER: So I think first and foremost is the parallel, if you will, to the predictions that you focused on in your book, which is this will transform a lot of the core data processing tasks, the information tasks. And sure, the doctors and nurses is one thing. But if you just think of clinical trial operations or the submission of regulatory documents, these are all kind of simple data … they’re not simple, obviously, but they’re data processing tasks. They involve natural language. That’s not going to be our focus, but I hope that others will use that to make clinical trials faster, more efficient, less expensive.
There’s already a lot of progress that’s happening on the molecular design side of things and taking hypotheses and turning them quickly and effectively into molecules. As I said, this is part of our work that we absolutely do and we don’t talk about it very much, simply because it’s a very crowded landscape and a lot of companies are engaged on that. But I think it’s really important to be able to take biological insights and turn them into new molecules.
And then, of course, the transformer models and their likes play a very significant role in that sort of turning insights into molecules because you can have foundation models for proteins. There are increasing efforts to create foundation models for other categories of molecules. And so that will undoubtedly accelerate the process by which you can quickly generate different molecular hypotheses and test them and learn from what you did so that you can do fewer iterations …
LEE: Right.
KOLLER: … before you converge on a successful molecule.
I do think that arguably the biggest impact as yet to be had is in that understanding of core human biology and what are the right ways to intervene in it. And that plays a role in a couple different ways. First of all, it certainly plays a role in which … if we are able to understand the human physiological state and, you know, the state of different systems all the way down to the cell level, that will inform our ability to pick hypotheses that are more likely to actually impact the right biologies underneath.
LEE: Yep. Yeah.
KOLLER: And the more data we’re able to collect about humans and about cells, the more successful our models will be at representing that human physiological state or the cell biological state and making predictions reliably on the impact of these interventions.
The other side of it, though, and this comes back, I think, to themes that were very much in your book, is this will impact not only the early stages of which hypotheses we interrogate, which molecules we move forward, but also hopefully at the end of the day, which molecule we prescribe to which patient.
LEE: Right.
KOLLER: And I think there’s been obviously so much narrative over the years about precision medicine, personalized medicine, and very little of that has come to fruition, with the exception of, you know, certain islands in oncology, primarily on genetically driven cancers.
But I think the opportunity is still there. We just haven’t been able to bring it to life because of the lack of the right kind of data. And I think with the increasing amount of human, kind of, foundational data that we’re able to acquire, things that are not sort of distilled through the eye of a clinician, for example, …
LEE: Yes.
KOLLER: … but really measurements of human pathology, we can start to get to some of that precision, carving out of the human population and then get to a world where we can prescribe the right medicine to the right patient and not only in cancer but also in other diseases that are also not a single disease.
LEE: All right, so now to wrap up this time together, I always try to ask one more provocative last question. One of the dreams that comes naturally to someone like me or any of my colleagues, probably even to you, is this idea of, you know, wouldn’t it be possible someday to have a foundation model for biology or for human biology or foundation model for the human cell or something along these lines?
And in fact, there are, of course, you and I are both aware of people who are taking that idea seriously and chasing after it. I have people in our labs that think hard about this kind of thing. Is it a reasonable thought at all?
KOLLER: I have learned over the years to avoid saying the word never because technology proceeds in ways that you often don’t expect. And so will we at some point be able to measure the cell in enough different ways across enough different channels at the same time that you can piece together what a cell does? I think that is eminently feasible, not today, but over time.
I don’t think it’s feasible using today’s technology, although the efforts to get there may expose where the biggest opportunities lie to, you know, build that next layer. So I think it’s good that people are working on really hard problems. I would also point out that even if one were to solve that really challenging problem of creating a model of a cell, there is thousands of different types of cells within the human body.
They’re very different. They also talk to each other …
LEE: Yep.
KOLLER: … both within the cell type and across different cell types. So the combinatorial complexity of that system is, I think, unfathomable to many people. I mean, I would say to all of us.
LEE: Yeah.
KOLLER: And so even from that very lofty goal, there is multiple big steps that would need to be taken to a mechanistic model of the full organism. So will we ever get there? Again, you know, I don’t see a reason why this is impossible to do. So I think over time, technology will get better and will allow us to build more and more elaborate models of more and more complex systems.
Patients can’t wait …
LEE: Right. Yeah.
KOLLER: … for that to happen in order for us to get them better medicines. So I think there is a great basic science initiative on that side of things. And, in parallel, we need to make do with the data that we have or can collect or can print. We print a lot of data in our internal wet labs and get to drugs that are effective even though they don’t benefit from having a full-blown mechanistic model.
LEE: Last question: where do you think we’ll be in five years?
KOLLER: Phew. If I had answered that question five years ago, I would have been very badly embarrassed at the inaccuracy of my answer. [LAUGHTER] So I will not answer it today either.
I will say that the thing about exponential curves is that they are very, very tricky, and they move in unexpected ways. I would hope that in five years, we will have made a sufficient investment in the generation of scientific data that we will be able to move beyond data that was generated entirely by humans and therefore insights that are derivative of what people already know to things that are truly novel discoveries.
And I think in order to do that in, you know, math, maybe because math is entirely conceptual, maybe you can do that today. Math is effectively a construct of the human mind. I don’t think biology is a construct of the human mind, and therefore one needs to collect enough data to really build those models that will give rise to those novel insights.
And that’s where I hope we will have made considerable progress in five years.
LEE: Well, I’m with you. I hope so, too. Well, you know, thank you, Daphne, so much for this conversation. I learn a lot talking to you, and it was great to, you know, connect again on this. And congratulations on all of this success. It’s really groundbreaking.
KOLLER: Thank you very much, Peter. It was a pleasure chatting with you, as well.
[TRANSITION MUSIC]
LEE: I still think of Daphne first and foremost as an AI researcher. And for sure, her research work in machine learning continues to be incredibly influential to this day. But it’s her work on AI-enhanced drug development that now is on the verge of making a really big difference on some of the most difficult diseases afflicting people today.
In our book, Carey, Zak, and I predicted that AI might be a meaningful accelerant in biomedical research, but I don’t know that we foresaw the incredible potential specifically in drug development.
Today, we’re seeing a flurry of activity at companies, universities, and startups on generative AI systems that aid and maybe even completely automate the design of new molecules as drug candidates. But now, in our conversation with Daphne, seeing AI go even further than that to do what one might reasonably have assumed to be impossible, to identify and select novel drug targets, especially for a neurodegenerative disease like ALS, it’s just, well, mind blowing.
Let’s continue our deep dive on AI and biomedical research with this conversation with Noubar Afeyan:
LEE: Noubar, thanks so much for joining. I’m really looking forward to this conversation.
NOUBAR AFEYAN: Peter, thanks. Thrilled to be here.
LEE: While I think most of the listeners to this podcast have heard of Flagship Pioneering (opens in new tab), it’s still worth hearing from you, you know, what is Flagship? And maybe a little bit about your background. And finally, you found a way to balance science and business creation. And so, you know, your approach and philosophy to all of that.
AFEYAN: Well, great. So maybe I’ll just start out by way of quick background. You know, my … and since we’re going talk about AI, I’ll also highlight my first contact with the topic of AI. So as an undergraduate in 1980 up at McGill University, I was an engineering student, but I was really captivated by, at that time, the talk on the campus around the expert system, heuristic-based, rule-based kind of programs.
LEE: Right.
AFEYAN: And so actually I had the dubious distinction of writing my one and only college newspaper article. [LAUGHTER] That was a short career. And it was all about how artificial intelligence would be impacting medicine, would be impacting, you know, speech capture, translation, and some of the ideas that were there that it’s interesting to see now 45 years later re-emerge with some of the new learning-based models.
My journey after college ended up taking me into biotechnology. In the early ’80s, I came to MIT to do a PhD. At the time, the field was brand new. I ended up being the first PhD graduate from MIT in this combination biology and engineering degree. And since then, I’ve basically been—so since 1987—a founder, a technologist in the space of biotechnology for human health and as well for planetary health.
And then in 1999/2000 formed what is now Flagship Pioneering, which essentially was an attempt to bring together the three elements of what we know are important in startups. That is scientific capital, human capital, and financial capital. Right now, startups get that from different places. The science in our fields mostly come from academia, research hospitals. The human capital comes from other startups …
LEE: Yeah.
AFEYAN: … or large companies or some academics leave. And then the financial capital is usually venture capital, but there’s also now more and more other deeper pockets of money.
What we thought was, what if all that existed in one entity and instead of having to convince each other how much they should believe the other if we just said, “Let’s use that power to go work on much further out things”? But in a way where nobody would believe it in the beginning, but we could give ourselves a little bit of time to do impactful big things.
Twenty-five years later, that’s the road we’ve stayed on.
LEE: OK. So let’s get into AI. Now, you know, what I’ve been asking guests is kind of an origin story. And there’s the origin story of contact with AI, you know, before the emergence of generative AI and afterwards. I don’t think there’s much of a point to asking you the pre-ChatGPT. But … so let’s focus on your first encounter with ChatGPT or generative AI. When did that happen, and what went through your head?
AFEYAN: Yeah. So, if you permit me, Peter, just for very briefly, let me actually say I had the interesting opportunity over the last 25 years to actually stay pretty close to the machine learning world …
LEE: Yeah. Yeah.
AFEYAN: … because one, as you well know, among the most prolific users of machine learning has been the bioinformatics computational biology world because it’s been so data rich that anything that can be done, people have thrown at these problems because unlike most other things, we’re not working on man-made data. We’re looking at data that comes from nature, the complexity of which far exceeds our ability to comprehend.
So you could imagine that any approach to statistically reduce complexity, get signal out of scant data—that’s a problem that’s been around.
The other place where I’ve been exposed to this, which I’m going to come back to because that’s where it first felt totally different to me, is that some 25 years ago, actually the very first company we started was a company that attempted to use evolutionary algorithms to essentially iteratively evolve consumer-packaged goods online. Literally, we tried to, you know, consider features of products as genes and create little genomes of them. And by recombination and mutation, we could create variety. And then we could get people through panels online—this was 2002/2003 timeframe—we could essentially get people through iterative cycles of voting to create a survival of the fittest. And that’s a company that was called Affinnova.
The reason I say that is that I knew that there’s a much better way to do this if only: one, you can generate variety …
LEE: Yeah.
AFEYAN: … without having to prespecify genes. We couldn’t do that before. And, two, which we’ve come back to nowadays, you can actually mimic how humans think about voting on things and just get rid of that element of it.
So then to your question of when does this kind of begin to feel different? So you could imagine that in biotechnology, you know, as an engineer by background, I always wanted to do CAD, and I picked the one field in which CAD doesn’t exist, which is biology. Computer-aided design is kind of a notional thing in that space. But boy, have we tried. For a long time, …
LEE: Yep.
AFEYAN: … people would try to do, you know, hidden Markov models of genomes to try to figure out what should be the next, you know, base that you may want to or where genes might be, etc. But the notion of generating in biology has been something we’ve tried for a while. And in the late teens, so kind of 2018, ’17, ’18, because we saw deep learning come along, and you could basically generate novelty with some of the deep learning models … and so we started asking, “Could you generate a protein basically by training a correspondence table, if you will, between protein structures and their underlying DNA sequence?” Not their protein sequence, but their DNA sequence.
LEE: Yeah.
AFEYAN: So that’s a big leap. So ’17/’18, we started this thing. It was called 56. It was FL56, Flagship Labs 56, our 56th project.
By the way, we started this parallel one called “57” that did it in a very different way. So one of them did pure black box model-building. The other one said, you know what, we don’t want to do the kind of … at that time, AlphaFold was in its very early embodiments. And we said, “Is there a way we could actually take little, you know, multi amino acid kind of almost grammars, if you will, a little piece, and then see if we could compose a protein that way?” So we were experimenting.
And what we found was that actually, if you show enough instances and you could train a transformer model—back in the day, that’s what we were using—you could actually, say, predict another sequence that should have the same activity as the first one.
LEE: Yeah.
AFEYAN: So we trained on green fluorescent proteins. Now, we’re talking about seven years ago. We trained on enzymes, and then we got to antibodies.
With antibodies, we started seeing that, boy, this could be a pretty big deal because it has big market impact. And we started bringing in some of the diffusion models that were beginning to come along at that time. And so we started getting much more excited. This was all done in a company that subsequently got renamed from FL56 to Generate:Biomedicines (opens in new tab), …
LEE: Yep, yep.
AFEYAN: … which is one of the leaders in protein design using the generative techniques. It was interesting because Generate:Biomedicines is a company that was called that before generative AI was a thing, [LAUGHTER] which was kind of very ironic.
And, of course, that team, which operates today very, very kind of at the cutting edge, has published their models. They came up with this first Chroma (opens in new tab) model, which is a diffusion-based model, and then started incorporating a lot of the LLM capabilities and fusing them.
Now we’re doing atomistic models and many other things. The point being, that gave us a glimpse of how quickly the capability was gaining, …
LEE: Yeah. Yeah.
AFEYAN: … just like evolution shows you. Sometimes evolution is super silent, and then all of a sudden, all hell breaks loose. And that’s what we saw.
LEE: Right. One of the things that I reflect on just in my own journey through this is there are other emotions that come up. One that was prominent for me early on was skepticism. Were there points when even in your own work, transformer-based work on this early on, that you had doubts or skepticism that these transformer architectures would be or diffusion-based approaches would be worth anything?
AFEYAN: You know, it’s interesting, I think that, I’m going to say this to you in a kind of a friendly way, but you’ll understand what I mean. In the world I live in, it’s kind of like the slums of innovation, [LAUGHTER] kind of like just doing things that are not supposed to work. The notion of skepticism is a luxury, right. I assume everything we do won’t work. And then once in a while I’m wrong.
And so I don’t actually try to evaluate whether before I bring something in, like just think about it. We, some hundred or so times a year, ask “what if” questions that lead us to totally weird places of thought. We then try to iterate, iterate, iterate to come up with something that’s testable. Then we go into a lab, and we test it.
So in that world, right, sitting there going, like, “How do I know this transformer is going to work?” The answer is, “For what?” Like, it’s going to work. To make something up … well, guess what? We knew early on with LLMs that hallucination was a feature, not a bug for what we wanted to do.
So it’s just such a different use that, of course, I have trained scientific skepticism, but it’s a little bit like looking at a competitive situation in an ecology and saying, “I bet that thing’s going to die.” Well, you’d be right—most of the time, you’d be right. [LAUGHTER]
So I just don’t … like, it … and that’s why—I guess, call me an early adopter—for us, things that could move the needle even a little, but then upon repetition a lot, let alone this, …
LEE: Yeah.
AFEYAN: … you have to embrace. You can’t wait there and say, I’ll embrace it once it’s ready. And so that’s what we did.
LEE: Hmm. All right. So let’s get into some specifics and what you are seeing either in your portfolio companies or in the research projects or out in the industry. What is going on today with respect to AI really being used for something meaningful in the design and development of drugs?
AFEYAN: In companies that are doing as diverse things as—let me give you a few examples—a project that’s now become a named company called ProFound Therapeutics (opens in new tab) that literally discovered three, four years ago, and would not have been able to without some of the big data-model-building capabilities, that our cells make literally thousands, if not tens of thousands, of more proteins than we were aware of, full stop.
We had done the human genome sequence, there was 20,000 genes, we thought that there was …
LEE: Wow.
AFEYAN: … maybe 70-80,000, 100,000 proteins, and that’s that. And it turns out that our cells have a penchant to express themselves in the form of proteins, and they have many other ways than we knew to do that.
Now, so what does that mean? That means that we have generated a massive amount of data, the interpretation of which, the use of which to guide what you do and what these things might be involved with is purely being done using the most cutting-edge data-trained models that allow you to navigate such complexity.
LEE: Wow. Hmm.
AFEYAN: That’s just one example. Another example: a company called Quotient Therapeutics (opens in new tab), again three, four years old. I can talk about the ones that are three, four years old because we’ve kind of gotten to a place where we’ve decided that it’s not going to fail yet, [LAUGHTER] so we can talk about it.
You know, we discovered—our team discovered—that in our cells, right, so we know that when we get cancer, our cells have genetic mutations in them or DNA mutations that are correlated and often causal to the hyperproliferative stages of cancer. But what we assume is that all the other cells in our body, pretty much, have one copy of their genes from our mom, one copy from our dad, and that’s that.
And when very precise deep sequencing came along, we always asked the question, “How much variation is there cell to cell?”
LEE: Right.
AFEYAN: And the answer was it’s kind of noise, random variation. Well, our team said, “Well, what if it’s not really that random?” because upon cell division cycles, there’s selection happening on these cells. And so not just in cancer but in liver cells, in muscle cells, in skin cells …
LEE: Oh, interesting.
AFEYAN: … can you imagine that there’s an evolutionary experiment that is favoring either compensatory mutations that are helping you avoid disease or disease-caused mutations that are gaining advantage as a way to understand the mechanism? Sure enough—I wouldn’t be telling you otherwise—with massive amount of single cell sequencing from individual patient samples, we’ve now discovered that the human genome is mutated on average in our bodies 10,000 times, like over every base, like, it’s huge numbers.
And we’re finding very interesting big signals come out of this massive amount of data. By the way, data of the sort that the human mind, if it tries to assign causal explanations to what’s happening …
LEE: Right.
AFEYAN: … is completely inadequate.
LEE: When you think about a language model, we’re learning from human language, and the totality of human language—at least relative to what we’re able to compute today in terms of constructing a model—the totality of human language is actually pretty limited. And in fact, you know, as is always written about in click-baity titles, you know, the big model builders are actually starting to run short.
AFEYAN: Running out, running out, yes. [LAUGHTER]
LEE: But one of the things that perplexes me and maybe even worries me—like these two examples—are generally in the realm of cellular biology and the complexity. Let’s just take the example of your company, ProFound. You know, the complexity of what’s going on and the potential genetic diversity is such that, can we ever have enough data? You know, because there just aren’t that many human beings. There just aren’t that many samples.
AFEYAN: Well, it depends on what you want to train, right. So if you want to train a de novo evolutionary model that could take you from bacteria to human mammalian cells and the like, there may not be—and I’m not an expert in that—but that’s a question that we often kind of think about.
But if you’re trying to train a … like you know what the proteins we know about, how they interact with pathways and disease mechanisms and the like. Now all of a sudden you find out that there’s a whole continent of them missing in your explanations. But there are things you can reason, in quotations, through analogy, functional analogy, sequence analogy, homology. So there’s a lot of things that we could do to essentially make use of this, even though you may not have the totality of data needed to, kind of, predict, based on a de novo sequence, exactly what it’s going to do.
So I agree with the comparison. But … but you’re right. The complexity is … just keep in mind, on average, a protein may be interacting with 50 to 100 other proteins.
LEE: Right.
AFEYAN: So if you find thousands of proteins, you’ve found a massive interaction space through which information is being processed in a living cell.
LEE: But do you find in your AI companies that access to data ends up being a key challenge? Or, you know, how central is that?
AFEYAN: Access to data is a key challenge for the companies we have that are trying to build just models. But that’s the minority of things we do. The majority of things we do is to actually co-develop the data and the models. And as you know well, because you guys, you know, have given us some ideas around this space, that, you know, you could generate data and then think about what you’re to do with it, which is the way biotech is operated with bioinformatics.
LEE: Right, right.
AFEYAN: Or you could generate bespoke data that is used to train the model that’s quite separate from what you would have done in the natural course of biology. So we’re doing much more of the latter of late, and I think that’ll continue. So, but these things are proliferating.
I mean, it’s hard to find a place where we’re not using this. And the “this” is any and all data-driven model building, generative, LLM-based, but also every other technique to make progress.
LEE: Sure. So now moving away from the straight biochemistry applications, what about AI in the process of building a business, of making investment decisions, of actually running an operation? What are you seeing there?
AFEYAN: So, well, you know, Moderna, which is a company that I’m quite proud of being a founder and chairman of, has adopted a significant, significant amount of AI embedded into their operations in all aspects: from the manufacturing, quality control, the clinical monitoring, the design—every aspect. And in fact, they’ve had a partnership that they’ve had for a little while here with OpenAI, and they’ve tried many different ways to stay at the cutting edge of that.
So we see that play out at some scale. That’s a 5,000-, 6,000-person organization, and what they’re doing is a good example of what early adopters would do, at least in our kind of biotechnology company.
But then, you know, in our space, I would say the efficiency impact is kind of no different, than, you know, anywhere else in academia you might adopt it or in other kinds of companies. But where I find it an interesting kind of maybe segue is the degree to which it may fundamentally change the way we think about how to do science, which is a whole other use, right?
LEE: Right.
AFEYAN: So it’s not an efficiency gain per se, although it’s maybe an effectiveness gain when it comes to science, but can you just fundamentally train models to generate hypotheses?
LEE: Yep.
AFEYAN: And we have done that, and we’ve been doing this for the last three years. And now it’s getting better and better, the better these reasoning engines are getting and kind of being able to extrapolate and train for novelty. Can you convert that to the world’s best experimental protocol to very precisely falsify your hypothesis, on and on?
That closing of that loop, kind of what we call autonomous science, which we’ve been trying to do for the last two, three years and are making some progress in, that to me is another kind of bespoke use of these things, not to generate molecules in its chemistry, but to change the behavior of how science is done.
LEE: Yeah. So I always end with a couple of provocative questions, but I need—before we do that, while we’re on this subject—to get your take on Lila Sciences (opens in new tab).
And there is a vision there that I think is very interesting. It’d be great to hear it described by you.
AFEYAN: Sure. So Lila, after operating for two to three years in kind of a preparatory kind of stealth mode, we’ve now had a little bit more visibility around, and essentially what we’re trying to do there is to create what we call automated science factories, and such a factory would essentially be able to take problems, either computationally specified or human-specified, and essentially do the experimental work in order to either make an optimization happen or enable something that just didn’t exist. And it’s really, at this point, we’ve shown proof of concept in narrow areas.
LEE: Yep.
AFEYAN: But it’s hard to say that if you can do this, you can’t do some other things, so we’re just expanding it that way. We don’t think we need a complete proof or complete demonstration of it for every aspect.
LEE: Right.
AFEYAN: So we’re just kind of being opportunistic. The idea for Lila is to partner with a number of companies. The good news is, within Flagship, there’s 48 of them. And so there’s a whole lot of them they can partner with to get their learning cycles. But eventually they want to be a real alternative to every time somebody has an idea, having to kind of go into a lab and manually do this.
I do want to say one thing we touched on, Peter, though, just on that front, which is …
LEE: Yep.
AFEYAN: … if you say, like, “What problem is this going to solve?” It’s several but an important one is just the flat-out human capacity to reason on this much data and this much complexity that is real. Because nature doesn’t try to abstract itself in a human understandable form.
LEE: Right. Yeah.
AFEYAN: In biology, since it’s kind of like progress happens through evolutionary kind of selections, the evidence of which [has] long been lost, and so therefore, you just see what you have, and then it has a behavior. I really do think that there’s something to be said, and I want to—just for your audience—lay out a provocative, at least, thought on all this, which Lila is a beginning embodiment of, which is that I really think that what’s going to happen over the next five, 10 years, even while we’re all fascinated with the impending arrival of AGI [artificial general intelligence] is really what I call poly-intelligence, which is the combination of human intelligence, machine intelligence, AI, and nature’s intelligence.
We’re all fascinated at the human-machine interface. We know the human-nature interface, but imagine the machine-nature interface—that is, actually letting loose a digital kind of information processing life form through the algorithms that are being developed and the commensurately complex, maybe much more complex. We’ll see. And so now the question becomes, what does the human do?
And we’re living in a world which is human dominated, which means the humans say, “If I don’t understand it, it’s not real, basically. And if I don’t understand it, I can’t regulate it.” And we’re going to have to make peace with the fact that we’re not going to be able to predictably affect things without necessarily understanding them the way we could if we just forced ourselves to only work on problems we can understand. And that world we’re not ready for at all.
LEE: Yeah. All right. So this one I predict is going to be a little harder for you because I think while you think about the future, you live very much in the present. But I’d like you to make some predictions about what the biotech and biopharmaceutical industries are going to be able to do two years from now, five years from now, 10 years from now.
AFEYAN: Yeah, well, it’s hard for me because you know my nature, which is that I think this is all emergent.
LEE: Right.
AFEYAN: And so I would be the conceit of predicting. So I would say with likelihood positive predictive value of less than 10%, I’m happy to answer your question. So I’m not trying to score high [LAUGHTER] because I really think that my job is to envision it, not to predict it. And that’s a little bit different, right?
LEE: Yeah, I actually was trying to pick what would be the hardest possible question I could ask you, [LAUGHTER] and this is what I came up with.
AFEYAN: Yeah, no, no, I’m kidding here. So now look, I think that we will cross this threshold of understandability. And of course you’re seeing that in a lot of LLM things today. And of course, people are trying to train for things that are explainers and all that whole, there’s a whole world of that. But I think at some point we’re going to have to kind of let go and get comfortable working on things that, you know …
I sometimes tell people, you know, and I’m not the first, but scientists and engineers are different, it’s said, in that engineers work on things that they don’t wait until they get a full understanding of before they work with them. Well, now scientists are going to have to get used to that, too, right?
LEE: Yeah. Yeah.
AFEYAN: Because insisting that it’s only valid if it’s understandable. So, I would say, look, I hope that the time … for example, I think major improvements will be made in patient selection. If we can test drugs on patients that are more synchronized as to the stage of their disease …
LEE: Yep.
AFEYAN: … I think the answer will be much better. We’re working on that. It’s a company called Etiome (opens in new tab), very, very early stage. It’s really beautiful data, very early data that shows that when we talk about MASH [metabolic dysfunction-associated steatohepatitis], liver disease, when we talk about Parkinson’s, there’s such a heterogeneity, not only of the subset type of the disease, but the stage of the disease, that this notion that you have stage one cancer, stage two cancer, again, nobody told nature there’s stages of that kind. It’s a continuum.
But if you can synchronize based on training, kind of, the ability to detect who are the patients that are in enough of a close proximity that should be treated so that the trial—much smaller a trial size—could give you a drug, then afterwards, you can prescribe it using these approaches.
Kind of we’re going to find that what we thought is one disease is more like 15 diseases. That’s bad news because we’re not going to be able to claim that we can treat everything which we can. It’s good news in that there’s going to be people who are going to start making much more specific solutions to things.
LEE: Right.
AFEYAN: So I can imagine that. I can imagine a generation of, kind of, students who are going to be able to play in this space without having 25 years of graduate education on the subject. So what is deemed knowledge sufficient to do creative things will change. I can go on and on, but I think all this is very close by and it’s very exciting.
LEE: Noubar, I just always have so much fun, and I learn really a lot. It’s high-density learning when I talk to you. And so I hope our listeners feel the same way. It’s something I really appreciate.
AFEYAN: Well, Peter, thanks for this. And I think your listeners know that if I was asking you questions, you would be answering them with equal if not more fascinating stuff. So, thanks for giving me the chance to do that today.
[TRANSITION MUSIC]
LEE: I’m always fascinated by Noubar’s perspectives on fundamental research and how it connects to human health and the building of successful companies. I see him as a classic “systems thinker,” and by that, I mean he builds impressive things like Flagship Pioneering itself, which he created as a kind of biomedical innovation system.
In our conversation, I was really struck by the fact that he’s been thinking about the potential impact of transformers—transformers being the fundamental building block of large language models—as far back as 2017, when the first paper on the attention mechanism in transformers was published by Google.
But, you know, it isn’t only about using AI to do things like understand and design molecules and antibodies faster. It’s interesting that he is also pushing really hard towards a future where AI might “close the loop” from hypothesis generation, to experiment design, to analysis, and so on.
Now, here’s my conversation with Dr. Eric Topol:
LEE: Eric, it’s really great to have you here.
ERIC TOPOL: Oh, Peter, I’m thrilled to be here with you here at Microsoft.
LEE: You’re a super famous person. Extremely well known to researchers even in computer science, as we have here at Microsoft Research.
But the question I’d like to ask is, how would you explain to your parents what you do every day?
TOPOL: [LAUGHS] That’s a good question. If I was just telling them I’m trying to come up with better ways to keep people healthy, that probably would be the easiest way to do it because if I ever got in deeper, I would lose them real quickly. They’re not around, but just thinking about what they could understand.
LEE: Right.
TOPOL: I think as long as they knew it was work centered on innovative paths to promoting and preserving human health, that would get to them, I think.
LEE: OK, so now, kind of the second topic, and then we let the conversation flow, is about origin stories with respect to AI. And with most of our guests, you know, I factor that into two pieces: the encounters with AI before ChatGPT and what we call generative AI and then the first contacts after.
And, of course, you have extensive contact with both now. But let’s start with how you got interested in machine learning and AI prior to ChatGPT. How did that happen?
TOPOL: Yeah, it was out of necessity. So back, you know, when I started at Scripps at the end of ’06, we started accumulating, you know, massive datasets. First, it was whole genomes. We did one of the early big cohorts of 1,400 people of healthy aging. We called the Wellderly whole genome sequence (opens in new tab).
And then we started big in the sensor world, and then we started saying, what are we going to do with all this data, with electronic health records and all those sensors? And now we got whole genomes.
And basically, what we were doing, we were in hoarding mode. We didn’t have a way to meaningfully analyze it.
LEE: Right.
TOPOL: You would read about how, you know, data is the new oil and, you know, gold and whatnot. But we just didn’t have a way to extract the juice. And even when we wanted to analyze genomes, it was incredibly laborious.
LEE: Yeah.
TOPOL: And we weren’t extracting a lot of the important information. So that’s why … not having any training in computer science, when I was doing the … about three years of work to do the book Deep Medicine, I started really, first auto-didactic about, you know, machine learning. And then I started contacting a lot of the real top people in the field and hanging out with them, and learning from them, getting their views as to, you know, where we are today, what models are coming in the future.
And then I said, “You know what? We are going to be able to fix this mess.” [LAUGHS] We’re going to get out of the hoarding phase, and we’re going to get into, you know, really making a difference.
So that’s when I embraced the future of AI. And I knew, you know, back—that was six years ago when it was published and probably eight or nine years ago when I was doing the research, and I knew that we weren’t there yet.
You know, at the time, we were seeing the image interpretation. That was kind of the early promise. But really, the models that were transformative, the transformer models, they were incubating back in 2017. So people knew something was brewing.
LEE: Right. Yes.
TOPOL: And everyone said we’re going to get there.
LEE: So then, ChatGPT comes out November of 2022; there’s GPT-4 in 2023, and now a lot has happened. Do you remember what your first encounter with that technology was?
TOPOL: Oh, sure. First, ChatGPT. You know, in the last days of November ’22, I was just blown away. I mean, I’m having a conversation. I’m having fun. And this is humanoid responding to me. I said, “What?” You know? So that was to me, a moment I’ll never forget. And so I knew that the world was, you know, at a very kind of momentous changing point.
Of course, knowing, too, that this is going to be built on, and built on quickly. Of course, I didn’t know how soon GPT-4 and all the others were going to come forward, but that was a wake-up call that the capabilities of AI had just made a humongous jump, which seemingly was all of a sudden, although I did know this had been percolating …
LEE: Right.
TOPOL: … you know, for what, at least five years, that, you know, it really was getting into its position to do this.
LEE: I know one of the things that was challenging psychologically and emotionally for me is, it made me rethink a lot of things that were going on in Microsoft Research in areas like causal reasoning, natural language processing, speech processing, and so on.
I’m imagining you must have had some emotional struggles too because you have this amazing book, Deep Medicine. Did you have to … did it go through your mind to rethink what you wrote in Deep Medicine in light of this or, or, you know, how did that feel?
TOPOL: It’s funny you ask that because in this one chapter I have on the virtual health coach, I wrote a whole bunch of scenarios …
LEE: Yeah.
TOPOL: … that were very kind of futuristic. You know, about how the AI interacts with the person’s health and schedules their appointment for this and their scan and tells them what lab tests they should tell their doctor to have, and, you know, all these things. And I sent a whole bunch of these, thinking that they were a little too far-fetched.
LEE: Yes.
TOPOL: And I sent them to my editor when I wrote the book, and he says, “Oh, these are great. You should put them all in.” [LAUGHTER] What I didn’t realize is they weren’t that, you know, they were all going to happen.
LEE: Yeah. They weren’t that far-fetched at all.
TOPOL: Not at all. If there’s one thing I’ve learned from all this, is our imagination isn’t big enough.
LEE: Yeah.
TOPOL: We think too small.
LEE: Now in our book that Carey, Zak, and I wrote, you know, we made, you know, we sort of guessed that GPT-4 might help biomedical researchers, but I don’t think that any of us had the thought in mind that the architecture around generative AI would be so directly applicable to, you know, say, protein structures or, you know, to clinical health records and so on.
And so a lot of that seems much more obvious today. But two years ago, it wasn’t. But we did guess that biomedical researchers would find this interesting and be helped along.
So as you reflect over the past two years, you know, do you have things that you think are very important, kind of, meaningful applications of generative AI in the kinds of research that Scripps does?
TOPOL: Yeah. I mean, I think for one, you pointed out how the term generative AI is a misnomer.
LEE: Yeah.
TOPOL: And so it really was prescient about how, you know, it had a pluripotent capability in every respect, you know, of editing and creating. So that was something that I think was telling us, an indicator that this is, you know, a lot bigger than how it’s being labeled. And our expectations can actually be more than what we had seen previously with the earlier version.
So I think what’s happened is that now, we keep jumping. It’s so quick that we can’t … you know, first we think, oh, well, we’ve gone into the agentic era, and then we could pass that with reasoning. [LAUGHTER] And, you know, we just can’t …
LEE: Right.
TOPOL: It’s just wild.
LEE: Yeah.
TOPOL: So I think so many of us now will put in prompts that will necessitate or ideally result in a not-immediate gratification, but rather one that requires, you know, quite a bit of combing through the corpus of knowledge …
LEE: Yeah.
TOPOL: … and getting, with all the citations, a report or a response. And I think now this has been a reset because to do that on our own, it takes, you know, many, many hours. And it’s usually incomplete.
But one of the things that was so different in the beginning was you would get the references from up to a year and a half previously.
LEE: Yep.
TOPOL: And that’s not good enough. [LAUGHS]
LEE: Right.
TOPOL: And now you get references, like, from the day before.
LEE: Yes. Yeah.
TOPOL: And so, you say, “Why would you do a regular search for anything when you could do something like this?”
LEE: Yeah.
TOPOL: And then, you know, the reasoning power. And a lot of people who are not using this enough still are talking about, “Well, there’s no reasoning.”
LEE: Yeah.
TOPOL: Which you dealt with really well in the book. But what, of course, you couldn’t have predicted is the new dimensions.
LEE: Right.
TOPOL: I think you nailed it with GPT-4. But it’s all these just, kind of, stepwise progressions that have been occurring because of the velocity that’s unprecedented. I just can’t believe it.
LEE: We were aware of the idea of multi-modality, but we didn’t appreciate, you know, what that would mean. Like AlphaFold (opens in new tab) [protein structure database], you know, the ability for AI to understand—or crystal structures—to really start understanding something more fundamental about biochemistry or medicinal chemistry.
I have to admit, when we wrote the book, we really had no idea.
TOPOL: Well, I feel the same way. I still today can’t get over it because the reason AlphaFold and Demis [Hassabis] and John Jumper [AlphaFold’s co-creators] were so successful is there was this protein databank.
LEE: Yes.
TOPOL: And it had been kept for decades. And so, they had the substrate to work with.
LEE: Right.
TOPOL: So, you say, “OK, we can do proteins.” But then how do you do everything else?
LEE: Right.
TOPOL: And so this whole, what I call, “large language of life model” work, which has gone into high gear like I’ve never seen.
LEE: Yeah.
TOPOL: You know, now to this holy grail of a virtual cell, and …
LEE: Yeah.
TOPOL: You know, it’s basically … it’s … it was inspired by proteins. But now it’s hitting on, you know, ligands and small molecules, cells. I mean, nothing is being held back here.
LEE: Yeah.
TOPOL: So how could anybody have predicted that?
LEE: Right.
TOPOL: I sure wouldn’t have thought it would be possible at this point.
LEE: Yeah. So just to challenge you, where do you think that is going to be two years from now? Five years from now? Ten years from now? Like, so you talk about a virtual cell. Is that achievable within 10 years, or is that still too far out?
TOPOL: No, I think within 10 years for sure. You know the group that got assembled that Steve Quake (opens in new tab) pulled together?
LEE: Right.
TOPOL: I think has 42 authors in a paper (opens in new tab) in Cell. The fact that he could get these 42 experts in life science and some in computer science to come together and all agree …
LEE: Yeah.
TOPOL: … that not only is this a worthy goal, but it’s actually going to be realized, that was impressive.
I challenged him about that. How did you get these people all to agree? So many of them were naysayers. And by the time the workshop finished, they were fully convinced. I think that what we’re seeing is so much progress happening so quickly. And then all the different models, you know, across DNA, RNA, and everything are just zooming forward.
LEE: Yeah.
TOPOL: And it’s just a matter of pulling this together. Now when we have that, and I think it could easily be well before a decade and possibly, you know, between the five- and 10-year mark—that’s just a guess—but then we’re moving into another era of life science because right now, you know, this whole buzz about drug discovery.
LEE: Yep.
TOPOL: It’s not… with the ability to do all these perturbations at a cellular level.
LEE: Right.
TOPOL: Or the cell of interest.
LEE: Yeah.
TOPOL: Or the cell-to-cell interactions or the intra-cell interaction. So once you nail that, yeah, it takes it to a kind of another predictive level that we haven’t really fathomed. So, yes, there’s going to be drug discovery that’s accelerated. But this would make that and also the underpinnings of diseases.
LEE: Yeah.
TOPOL: So the idea that there’s so many diseases we don’t understand now. And if you had virtual cell, …
LEE: Yeah.
TOPOL: … you would probably get to that answer …
LEE: Yeah.
TOPOL: … much more quickly. So whether it’s underpinnings of diseases or what it’s going to take to really come up with far better treatments—preventions—I think that’s where virtual cell will get us.
LEE: There’s a technical question … I wonder if you have an opinion. You may or may not. There is sort of what I would refer to as ab initio approaches to this. You know, you start from the fundamental physics and chemistry, and we know the laws, we have the math and, you know, we can try to derive from there … in fact, we can even run simulations of that math to generate training data to build generative models and work up to a cell, or forget all of that and just take as many observations and measurements of, say, living cells as possible, and just have faith that hidden amongst all of the observational data, there is structure and language that can be derived.
So that’s sort of bottom-up versus top-down approaches. Do you have an opinion about which way?
TOPOL: Oh, I think you go after both. And clearly whenever you’re positing that you’ve got a virtual cell model that’s working, you’ve got to do the traditional methods as well to validate it, and … so all that. You know, I think if you’re going to go out after this seriously, you have to pull out all the stops. Both approaches, I think, are going to be essential.
LEE: You know, if what you’re saying is true, and it is amazing to hear the confidence, the one thing I tried to explain to someone nontechnical is that for a lot of problems in medicine, we just don’t have enough data in a really profound way. And the most profound way to say that is, since Adam and Eve, there have only been an estimated 106 billion people who have ever lived.
So even if we had the DNA of every human being, every individual of Homo sapiens, there are certain problems for which we would not have enough data.
TOPOL: Sure.
LEE: And so I think another thing that seems profound to me, if we can actually have a virtual cell, is we can actually make trillions of virtual …
TOPOL: Yeah
LEE: … human beings. The true genetic diversity could be realized for our species.
TOPOL: I think you nailed it. The ability to have that type of data, no less synthetic data, I mean, it’s just extraordinary.
LEE: Yeah.
TOPOL: We will get there someday. I’m confident of that. We may be wrong in projections. And I do think [science writer] Philip Ball won’t be right that it will never happen, though. [LAUGHTER] No, I think that if there’s a holy grail of biology, this is it.
LEE: Yeah.
TOPOL: And I think you’re absolutely right about where that will get us.
LEE: Yeah.
TOPOL: Transcending the beginning of the species.
LEE: Yeah.
TOPOL: Of our species.
LEE: Yeah. All right. So now, we’re starting to run short on time here. And so I wanted to ask you about, I’m in my 60s, so I actually think about this a lot more. [LAUGHTER] And I know you’ve been thinking a lot about longevity. And, of course, your new book, Super Agers.
And one of the reasons I’m so eager to read is it’s a topic very top of mind for me and actually for a lot of people. Where is this going? Because this is another area where you hear so much hype. At the same time, you see Nobel laureate scientists …
TOPOL: Yeah.
LEE: … working on this.
TOPOL: Yeah.
LEE: So, so what’s, what’s real there?
TOPOL: Yeah. Well, it’s really … the real deal is the science of aging is zooming forward.
And that’s exciting. But I see it bifurcating. On the one hand, all these new ideas, strategies to reverse aging are very ambitious. Like cell reprogramming and senolytics and, you know, the rejuvenation of our thymus gland, and it’s a long list.
LEE: Yeah.
TOPOL: And they’re really cool science, and it used to be the mouse lived longer. Now it’s the old mouse looks really young.
LEE: Yeah. Yeah.
TOPOL: All the different features. A blind mouse with cataracts is all of a sudden there’s no cataracts. I mean, so these things are exciting, but none of them are proven in people, and they all have significant risk, no less, you know, the expense that might be attached.
LEE: Right.
TOPOL: And some people are jumping the gun. They’re taking rapamycin, which can really knock out their immune system. So they all carry a lot of risk. And people are just getting a little carried away. We’re not there yet.
But the other side, which is what I emphasize in the book, which is exciting, is that we have all these new metrics that came out of the science of aging.
LEE: Yes.
TOPOL: So we have clocks of the body. Our biological clock versus our chronological clock, and we have organ clocks. So I can say, you know, Peter, we’ve assessed all your organs and your immune system. And guess what? Every one of them is either at or less than your actual age.
LEE: Right.
TOPOL: And that’s very reassuring. And by the way, your methylation clock is also … I don’t need to worry about you so much. And then I have these other tests that I can do now, like, for example, the brain. We have an amazing protein p-Tau217 that we can say over 20 years in advance of you developing Alzheimer’s, …
LEE: Yeah.
TOPOL: … we can look at that, and it’s modifiable by lifestyle, bringing it down. It should be you can change the natural history. So what we’ve seen is an explosion of knowledge of metrics, proteins, no less, you know, our understanding at the gene level, the gut microbiome, the immune system. So that’s what’s so exciting. How our immune system ages. Immunosenescence. How we have more inflammation—inflammaging—with aging. So basically, we have three diseases that kill us, that take away our health: heart, cancer, and neurodegenerative.
LEE: Yep.
TOPOL: And they all take more than 20 years. They all have a defective immune system inflammation problem, and they’re all going to be preventable.
LEE: Yeah.
TOPOL: That’s what’s so exciting. So we don’t have to have reverse aging. We can actually work on …
LEE: Just prevent aging in the first place.
TOPOL: … the age-related diseases. So basically, what it means is: I got to find out if you have a risk, if you’re in this high-risk group for this particular condition, because if you are—and we have many levels, layers, orthogonal ways to check—we don’t just bank it all on one polygenic test. We’re going to have several ways, say this is the one we are going …
And then we go into high surveillance, where, let’s say if it’s your brain, we do more p-Tau, if we need to do brain imaging—whatever it takes. And also, we do preventive treatments on top of the lifestyle [changes], that one of the problems we have today is a lot of people know generally, what are good lifestyle factors. Although, I go through a lot more than people generally acknowledge.
But they don’t incorporate them because they don’t know that they’re at risk and they could change their … extend their health span and prevent that disease. So what I at least put out there, a blueprint, is how we can use AI, because it’s multimodal AI, with all these layers of data, and then temporally, it’s like today you could say if you have two protein tests, not only are you going to have Alzheimer’s, but within a two-year time frame when …
LEE: Yep.
TOPOL: … and if you don’t change things, if we don’t gear up … you know, we can … we can completely prevent this, so … or at least defer it for a decade or more. So that’s why I’m excited, is that we made these strides in the science of aging. But we haven’t acknowledged the part that doesn’t require reversing aging. There’s this much less flashy, attainable, less risky approach …
LEE: Yeah.
TOPOL: … than the one that … when you reverse aging, you’re playing with the hallmarks of cancer. They are like, if you look at the hallmarks of cancer …
LEE: That has been one of the primary challenges.
TOPOL: They’re lined up.
LEE: Yeah.
TOPOL: They’re all the same, you know, whether it’s telomeres, or whether it’s … you know … so this is the problem. I actually say in the book, I do think one of these—we have so many shots on goal—one of these reverse aging things will likely happen someday. But we’re nowhere close.
On the other hand, let’s gear up. Let’s do what we can do. Because we have these new metrics that’s … people don’t … like, when I read the organ clock paper (opens in new tab) from Tony Wyss-Coray from Stanford. It was published end of ’23; it was the cover of Nature. It blew me away.
LEE: Yeah.
TOPOL: And I wrote a Substack (opens in new tab) [article] on it. And Tony said, “Well, that’s so nice of you.” I said, “So nice? This is revolutionary, you know.” [LAUGHTER] So …
LEE: By the way, what’s so interesting is, how these things, this kind of understanding and AI, are coming together.
TOPOL: Yes.
LEE: It’s almost eerie the timing of these things.
TOPOL: Absolutely. Because you couldn’t take all these layers of data, just like we were talking about data hoarding.
LEE: Yep.
TOPOL: Now we have data hoarding on individual with no way to be able to make these assessments of what level of risk, when, what are we going to do in this individual to prevent that? We can do that now.
We can do it today. And we could keep building on that. So I’m really excited about it. I think that, you know, when I wrote the last book on deep medicine, it was our overarching goal should be to bring back the patient-doctor relationship. I’m an old dog, and I know what it used to be when I got out of medical school.
It’s totally … you couldn’t imagine how much erosion from the ’70s, ’80s to now. But now I have a new overarching goal. I’m thinking that that still is really important—humanity in medicine—but let’s prevent these three … big three diseases because it’s an opportunity that we’re not … you know, in medicine, all my life we’ve been hearing and talking about we need to prevent diseases.
Curing is much harder than prevention. And the economics. Oh my gosh. But we haven’t done it.
LEE: Yeah.
TOPOL: Now we can do it. Primary prevention. We’d do really well. Somebody’s had heart attack.
LEE: Yeah.
TOPOL: Oh, we’re going to get all over it. Why did they have a heart attack in the first place?
LEE: Well, the thing that makes so much sense in what you’re saying is that we understand we have an understanding both economically and medically that prevention is a good thing. And extending the concept of prevention to these age-related conditions, I think, makes all the sense in the world.
You know, Eric, maybe on that optimistic note, it’s time to wrap up this conversation. Really appreciate you coming. Let me just brag in closing that I’m now the proud owner of an autographed copy of your latest book, and, really, thank you for that.
TOPOL: Oh, thank you. I could spend the rest of the day talking to you. I’ve really enjoyed it. Thanks.
[TRANSITION MUSIC]
LEE: For me, the biggest takeaway from our conversation was Eric’s supremely optimistic predictions about what AI will allow us to do in much less than 10 years.
You know, for me personally, I started off several years ago with the typical techie naivete that if we could solve protein folding using machine learning, we would solve human biology. But as I’ve gotten smarter, I’ve realized that things are way, way more complicated than that, and so hearing Eric’s techno-optimism on this is really both heartening and so interesting.
Another thing that really caught my attention are Eric’s views on AI in medical diagnosis. That really stood out to me because within our labs here at Microsoft Research, we have been doing a lot of work on this, for example in creating foundation models for whole-slide digital pathology.
The bottom line, though, is that biomedical research and development is really changing and changing quickly. It’s something that we thought about and wrote briefly about in our book, but just hearing it from these three people gives me reason to believe that this is going to create tremendous benefits in the diagnosis and treatment of disease.
And in fact, I wonder now how regulators, such as the Food and Drug Administration here in the United States, will be able to keep up with what might become a really big increase in the number of animal and human studies that need to be approved. On this point, it’s clear that the FDA and other regulators will need to use AI to help process the likely rise in the pace of discovery and experimentation. And so stay tuned for more information about that.
[THEME MUSIC]
I’d like to thank Daphne, Noubar, and Eric again for their time and insights. And to our listeners, thank you for joining us. There are several episodes left in the series, including discussions on medical students’ experiences with AI and AI’s influence on the operation of health systems and public health departments. We hope you’ll continue to tune in.
Until next time.
[MUSIC FADES]





 Vivek Gangasani is a Worldwide Lead GenAI Specialist Solutions Architect for SageMaker Inference. He drives Go-to-Market (GTM) and Outbound Product strategy for SageMaker Inference. He also helps enterprises and startups deploy, manage, and scale their GenAI models with SageMaker and GPUs. Currently, he is focused on developing strategies and content for optimizing inference performance and GPU efficiency for hosting Large Language Models. In his free time, Vivek enjoys hiking, watching movies, and trying different cuisines.
Vivek Gangasani is a Worldwide Lead GenAI Specialist Solutions Architect for SageMaker Inference. He drives Go-to-Market (GTM) and Outbound Product strategy for SageMaker Inference. He also helps enterprises and startups deploy, manage, and scale their GenAI models with SageMaker and GPUs. Currently, he is focused on developing strategies and content for optimizing inference performance and GPU efficiency for hosting Large Language Models. In his free time, Vivek enjoys hiking, watching movies, and trying different cuisines. Kareem Syed-Mohammed is a Product Manager at AWS. He is focuses on enabling Gen AI model development and governance on SageMaker HyperPod. Prior to this, at Amazon QuickSight, he led embedded analytics, and developer experience. In addition to QuickSight, he has been with AWS Marketplace and Amazon retail as a Product Manager. Kareem started his career as a developer for call center technologies, Local Expert and Ads for Expedia, and management consultant at McKinsey.
Kareem Syed-Mohammed is a Product Manager at AWS. He is focuses on enabling Gen AI model development and governance on SageMaker HyperPod. Prior to this, at Amazon QuickSight, he led embedded analytics, and developer experience. In addition to QuickSight, he has been with AWS Marketplace and Amazon retail as a Product Manager. Kareem started his career as a developer for call center technologies, Local Expert and Ads for Expedia, and management consultant at McKinsey. Piyush Daftary is a Senior Software Engineer at AWS, working on Amazon SageMaker. His interests include databases, search, machine learning, and AI. He currently focuses on building performant, scalable inference systems for large language models. Outside of work, he enjoys traveling, hiking, and spending time with family.
Piyush Daftary is a Senior Software Engineer at AWS, working on Amazon SageMaker. His interests include databases, search, machine learning, and AI. He currently focuses on building performant, scalable inference systems for large language models. Outside of work, he enjoys traveling, hiking, and spending time with family. Chaitanya Hazarey leads software development for inference on SageMaker HyperPod at Amazon, bringing extensive expertise in full-stack engineering, ML/AI, and data science. As a passionate advocate for responsible AI development, he combines technical leadership with a deep commitment to advancing AI capabilities while maintaining ethical considerations. His comprehensive understanding of modern product development drives innovation in machine learning infrastructure.
Chaitanya Hazarey leads software development for inference on SageMaker HyperPod at Amazon, bringing extensive expertise in full-stack engineering, ML/AI, and data science. As a passionate advocate for responsible AI development, he combines technical leadership with a deep commitment to advancing AI capabilities while maintaining ethical considerations. His comprehensive understanding of modern product development drives innovation in machine learning infrastructure. Andrew Smith is a Senior Cloud Support Engineer in the SageMaker, Vision & Other team at AWS, based in Sydney, Australia. He supports customers using many AI/ML services on AWS with expertise in working with Amazon SageMaker. Outside of work, he enjoys spending time with friends and family as well as learning about different technologies.
Andrew Smith is a Senior Cloud Support Engineer in the SageMaker, Vision & Other team at AWS, based in Sydney, Australia. He supports customers using many AI/ML services on AWS with expertise in working with Amazon SageMaker. Outside of work, he enjoys spending time with friends and family as well as learning about different technologies.
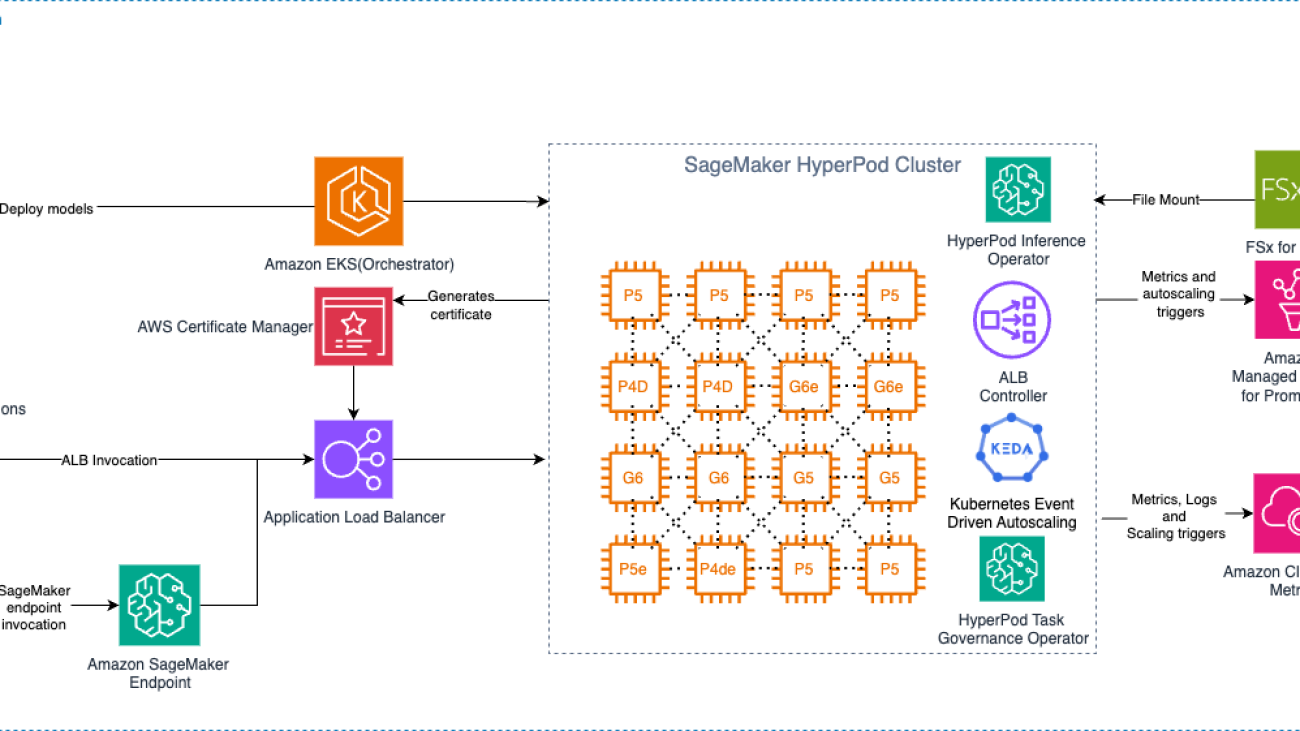
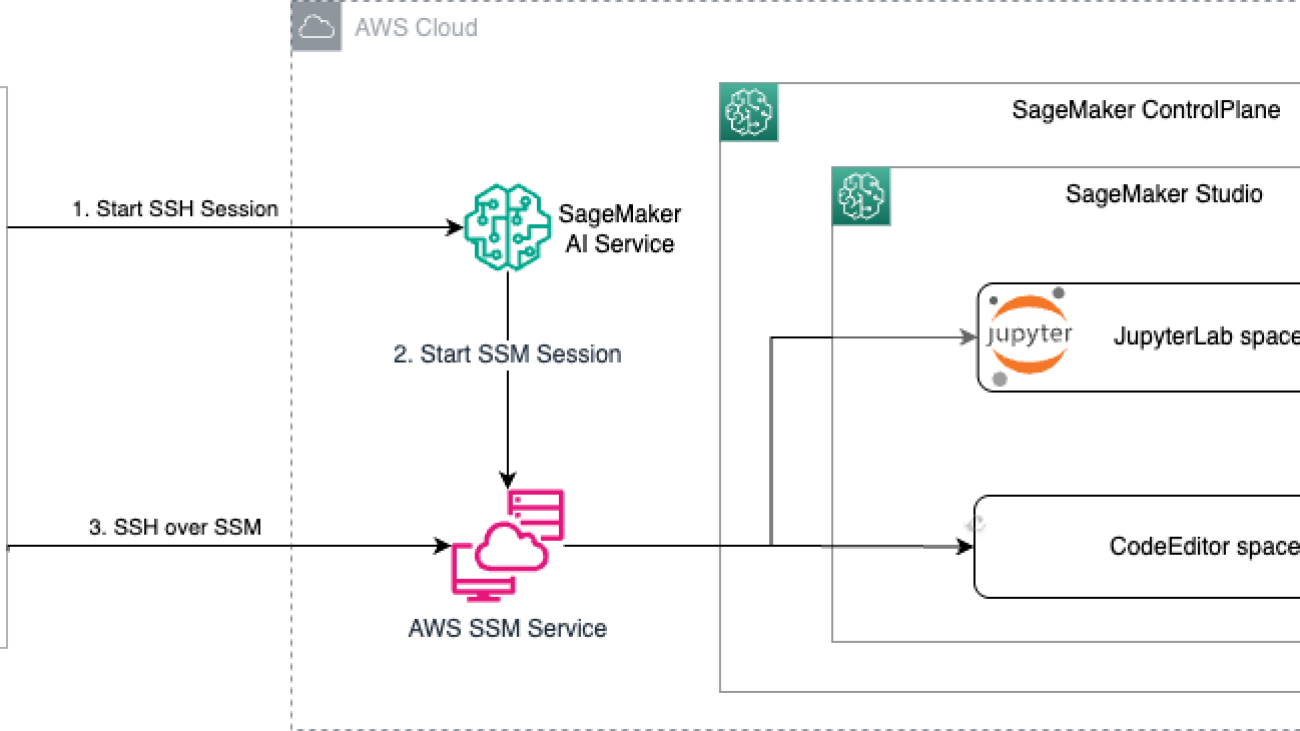











 Durga Sury is a Senior Solutions Architect at Amazon SageMaker, where she helps enterprise customers build secure and scalable AI/ML systems. When she’s not architecting solutions, you can find her enjoying sunny walks with her dog, immersing herself in murder mystery books, or catching up on her favorite Netflix shows.
Durga Sury is a Senior Solutions Architect at Amazon SageMaker, where she helps enterprise customers build secure and scalable AI/ML systems. When she’s not architecting solutions, you can find her enjoying sunny walks with her dog, immersing herself in murder mystery books, or catching up on her favorite Netflix shows. Edward Sun is a Senior SDE working for SageMaker Studio at Amazon Web Services. He is focused on building interactive ML solution and simplifying the customer experience to integrate SageMaker Studio with popular technologies in data engineering and ML landscape. In his spare time, Edward is big fan of camping, hiking, and fishing, and enjoys spending time with his family.
Edward Sun is a Senior SDE working for SageMaker Studio at Amazon Web Services. He is focused on building interactive ML solution and simplifying the customer experience to integrate SageMaker Studio with popular technologies in data engineering and ML landscape. In his spare time, Edward is big fan of camping, hiking, and fishing, and enjoys spending time with his family. Raj Bagwe is a Senior Solutions Architect at Amazon Web Services, based in San Francisco, California. With over 6 years at AWS, he helps customers navigate complex technological challenges and specializes in Cloud Architecture, Security and Migrations. In his spare time, he coaches a robotics team and plays volleyball. He can be reached at X handle @rajesh_bagwe.
Raj Bagwe is a Senior Solutions Architect at Amazon Web Services, based in San Francisco, California. With over 6 years at AWS, he helps customers navigate complex technological challenges and specializes in Cloud Architecture, Security and Migrations. In his spare time, he coaches a robotics team and plays volleyball. He can be reached at X handle @rajesh_bagwe. Sri Aakash Mandavilli is a Software Engineer on the Amazon SageMaker Studio team, where he has been building innovative products since 2021. He specializes in developing various solutions across the Studio service to enhance the machine learning development experience. Outside of work, SriAakash enjoys staying active through hiking, biking, and taking long walks.
Sri Aakash Mandavilli is a Software Engineer on the Amazon SageMaker Studio team, where he has been building innovative products since 2021. He specializes in developing various solutions across the Studio service to enhance the machine learning development experience. Outside of work, SriAakash enjoys staying active through hiking, biking, and taking long walks.

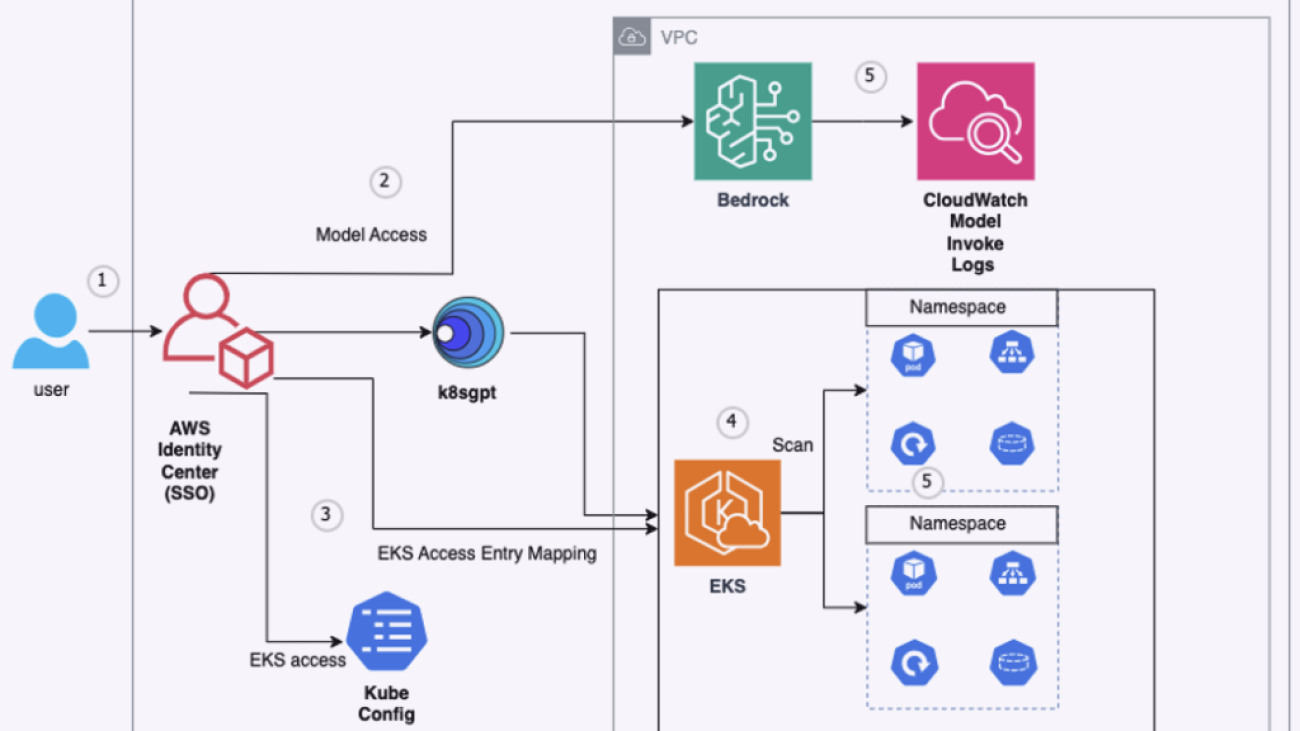







 Angela Wang is a Technical Account Manager based in Australia with over 10 years of IT experience, specializing in cloud-native technologies and Kubernetes. She works closely with customers to troubleshoot complex issues, optimize platform performance, and implement best practices for cost optimized, reliable and scalable cloud-native environments. Her hands-on expertise and strategic guidance make her a trusted partner in navigating modern infrastructure challenges.
Angela Wang is a Technical Account Manager based in Australia with over 10 years of IT experience, specializing in cloud-native technologies and Kubernetes. She works closely with customers to troubleshoot complex issues, optimize platform performance, and implement best practices for cost optimized, reliable and scalable cloud-native environments. Her hands-on expertise and strategic guidance make her a trusted partner in navigating modern infrastructure challenges. Haofei Feng is a Senior Cloud Architect at AWS with over 18 years of expertise in DevOps, IT Infrastructure, Data Analytics, and AI. He specializes in guiding organizations through cloud transformation and generative AI initiatives, designing scalable and secure GenAI solutions on AWS. Based in Sydney, Australia, when not architecting solutions for clients, he cherishes time with his family and Border Collies.
Haofei Feng is a Senior Cloud Architect at AWS with over 18 years of expertise in DevOps, IT Infrastructure, Data Analytics, and AI. He specializes in guiding organizations through cloud transformation and generative AI initiatives, designing scalable and secure GenAI solutions on AWS. Based in Sydney, Australia, when not architecting solutions for clients, he cherishes time with his family and Border Collies. Eva Li is a Technical Account Manager at AWS located in Australia with over 10 years of experience in the IT industry. Specializing in IT infrastructure, cloud architecture and Kubernetes, she guides enterprise customers to navigate their cloud transformation journeys and optimize their AWS environments. Her expertise in cloud architecture, containerization, and infrastructure automation helps organizations bridge the gap between business objectives and technical implementation. Outside of work, she enjoys yoga and exploring Australia’s bush walking trails with friends.
Eva Li is a Technical Account Manager at AWS located in Australia with over 10 years of experience in the IT industry. Specializing in IT infrastructure, cloud architecture and Kubernetes, she guides enterprise customers to navigate their cloud transformation journeys and optimize their AWS environments. Her expertise in cloud architecture, containerization, and infrastructure automation helps organizations bridge the gap between business objectives and technical implementation. Outside of work, she enjoys yoga and exploring Australia’s bush walking trails with friends. Alex Jones is a Principal Engineer at AWS. His career has focused largely on highly constrained environments for physical and digital infrastructure. Working at companies such as Microsoft, Canoncial and American Express, he has been both an engineering leader and individual contributor. Outside of work he has founded several popular projects such as OpenFeature and more recently the GenAI accelerator for Kubernetes, K8sGPT. Based in London, Alex has a partner and two children.
Alex Jones is a Principal Engineer at AWS. His career has focused largely on highly constrained environments for physical and digital infrastructure. Working at companies such as Microsoft, Canoncial and American Express, he has been both an engineering leader and individual contributor. Outside of work he has founded several popular projects such as OpenFeature and more recently the GenAI accelerator for Kubernetes, K8sGPT. Based in London, Alex has a partner and two children.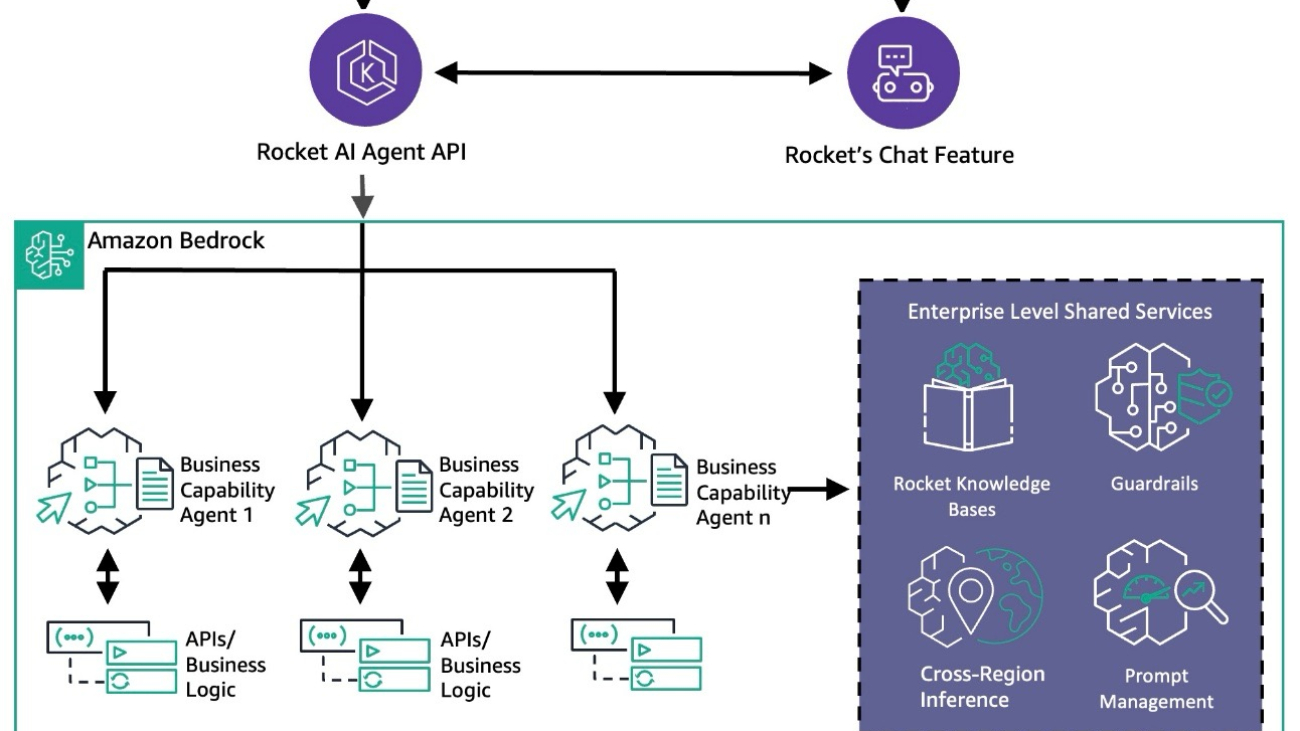

 Manali Sapre is a Senior Director at Rocket Mortgage, bringing over 20 years of experience leading transformative technology initiatives across the company. She has been at the forefront of innovation—spearheading Rocket’s first-generation AI chat platform, building the company’s original digital mortgage application, and launching scalable lead generation systems. Manali has also led multiple AI-driven initiatives focused on banker efficiency and internal productivity, helping to embed smart, human-centric technology into the daily workflows of team members. Her passion lies in solving complex challenges through collaboration, mentoring the next generation of tech leaders, and creating intuitive, high-impact experiences. Outside of work, Manali enjoys hiking, traveling, and spending quality time with her family.
Manali Sapre is a Senior Director at Rocket Mortgage, bringing over 20 years of experience leading transformative technology initiatives across the company. She has been at the forefront of innovation—spearheading Rocket’s first-generation AI chat platform, building the company’s original digital mortgage application, and launching scalable lead generation systems. Manali has also led multiple AI-driven initiatives focused on banker efficiency and internal productivity, helping to embed smart, human-centric technology into the daily workflows of team members. Her passion lies in solving complex challenges through collaboration, mentoring the next generation of tech leaders, and creating intuitive, high-impact experiences. Outside of work, Manali enjoys hiking, traveling, and spending quality time with her family. Seshidhar Raghupathi is a software architect at Rocket with over 12 years of experience driving innovation, scalability, and system resilience across AI and client communication platforms. He was instrumental in developing Rocket’s first cloud-based digital mortgage application and has since led several impactful initiatives to enhance intelligent, personalized client experiences. His expertise spans backend architecture, AI integration, platform modernization, and cross-team enablement. He is known for his ability to execute tactically while aligning with long-term strategic goals, particularly in enhancing security, scalability, and user experience. Outside of work, Seshi enjoys spending time with family, playing sports, and connecting with friends.
Seshidhar Raghupathi is a software architect at Rocket with over 12 years of experience driving innovation, scalability, and system resilience across AI and client communication platforms. He was instrumental in developing Rocket’s first cloud-based digital mortgage application and has since led several impactful initiatives to enhance intelligent, personalized client experiences. His expertise spans backend architecture, AI integration, platform modernization, and cross-team enablement. He is known for his ability to execute tactically while aligning with long-term strategic goals, particularly in enhancing security, scalability, and user experience. Outside of work, Seshi enjoys spending time with family, playing sports, and connecting with friends. Venkata Santosh Sajjan Alla is a Senior Solutions Architect at AWS Financial Services, driving AI-led transformation across North America’s FinTech sector. He partners with organizations to design and execute cloud and AI strategies that speed up innovation and deliver measurable business impact. His work has consistently translated into millions in value through enhanced efficiency and additional revenue streams. With deep expertise in AI/ML, Generative AI, and cloud-native architectures, Sajjan enables financial institutions to achieve scalable, data-driven outcomes. When not architecting the future of finance, he enjoys traveling and spending time with family. Connect with him on
Venkata Santosh Sajjan Alla is a Senior Solutions Architect at AWS Financial Services, driving AI-led transformation across North America’s FinTech sector. He partners with organizations to design and execute cloud and AI strategies that speed up innovation and deliver measurable business impact. His work has consistently translated into millions in value through enhanced efficiency and additional revenue streams. With deep expertise in AI/ML, Generative AI, and cloud-native architectures, Sajjan enables financial institutions to achieve scalable, data-driven outcomes. When not architecting the future of finance, he enjoys traveling and spending time with family. Connect with him on  Axel Larsson is a Principal Solutions Architect at AWS based in the greater New York City area. He supports FinTech customers and is passionate about helping them transform their business through cloud and AI technology. Outside of work, he is an avid tinkerer and enjoys experimenting with home automation.
Axel Larsson is a Principal Solutions Architect at AWS based in the greater New York City area. He supports FinTech customers and is passionate about helping them transform their business through cloud and AI technology. Outside of work, he is an avid tinkerer and enjoys experimenting with home automation.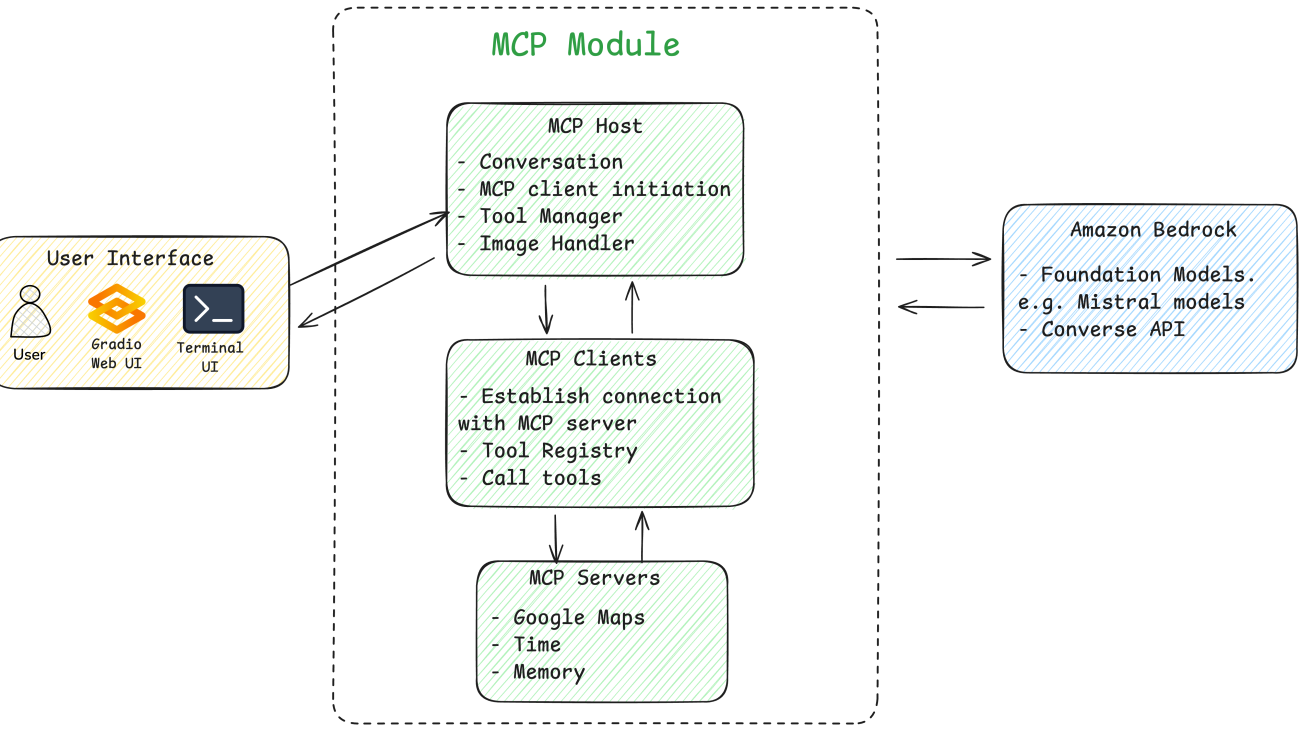
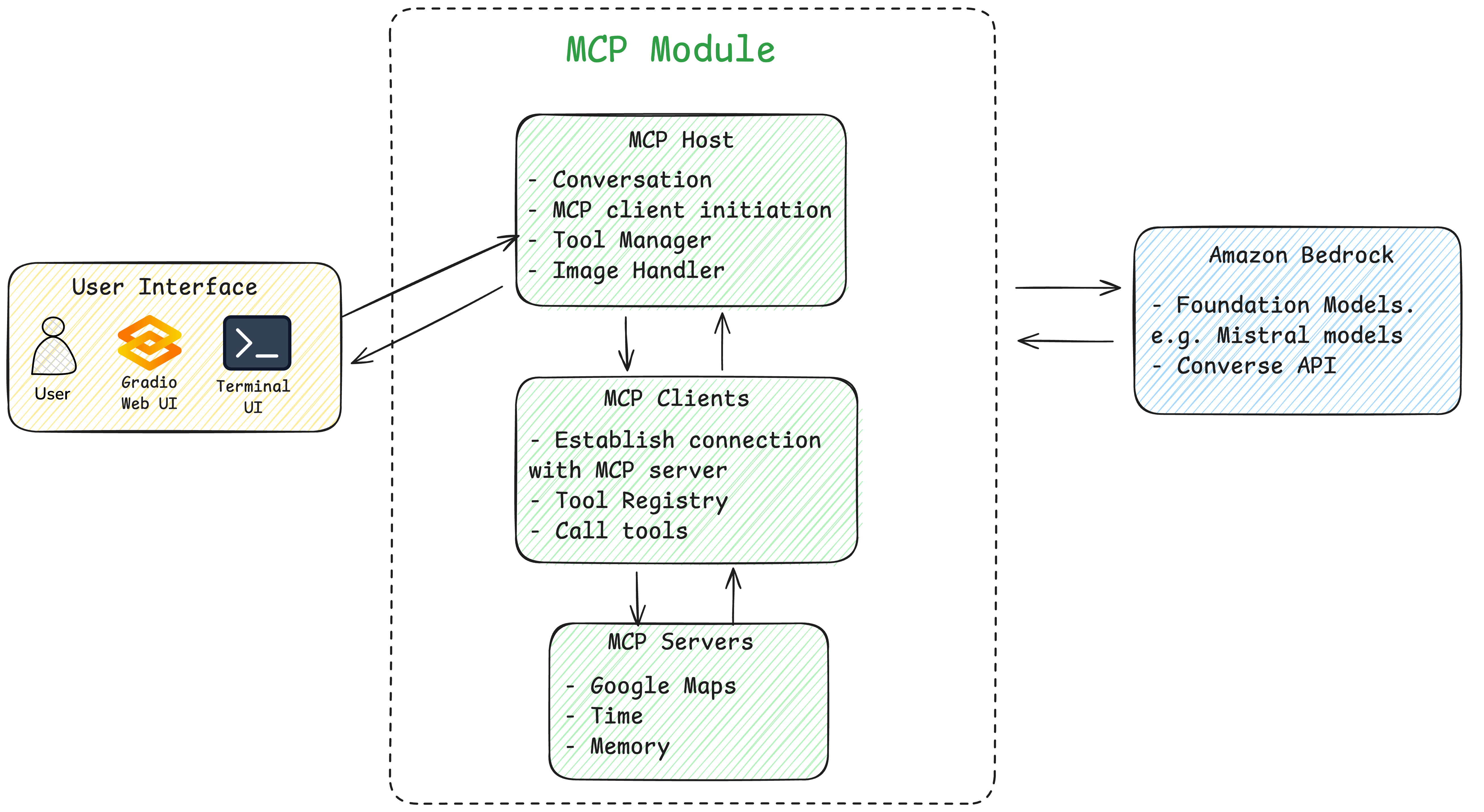
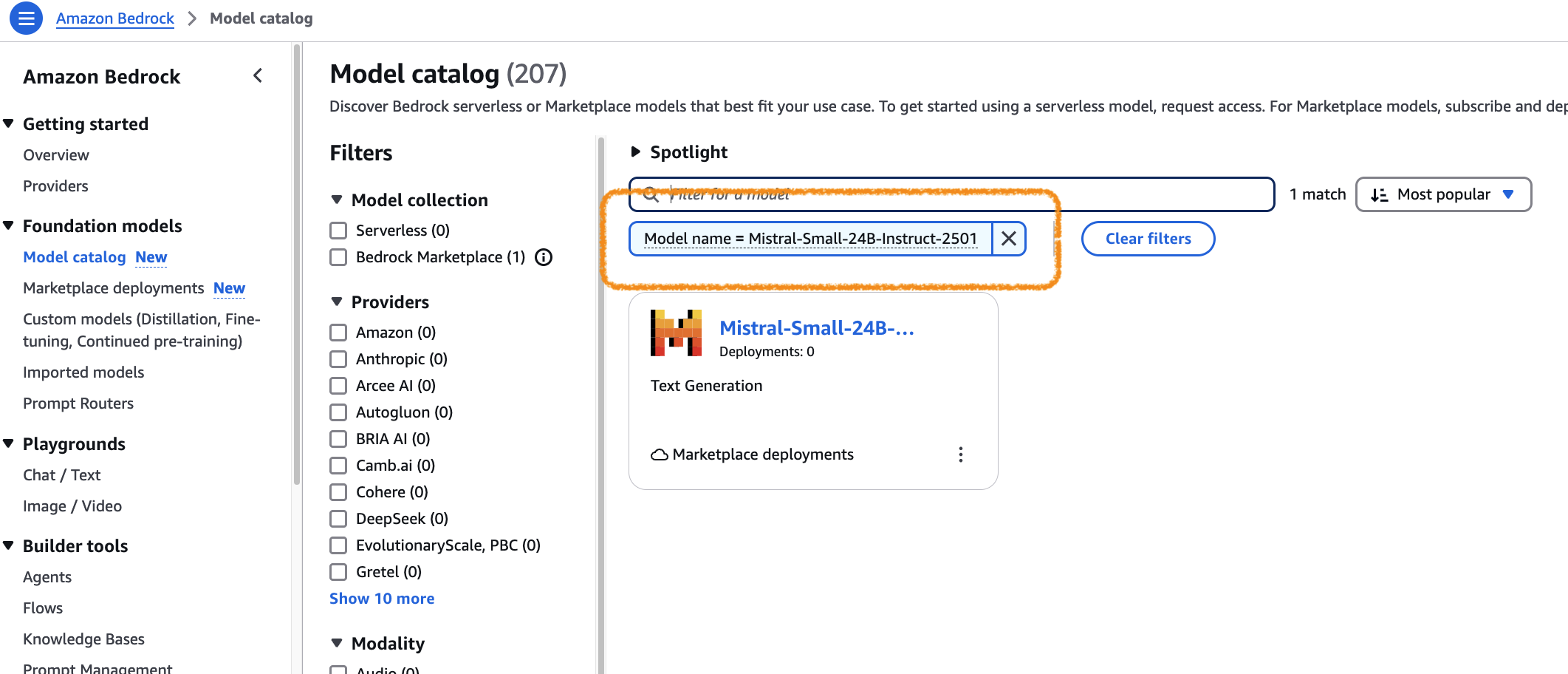
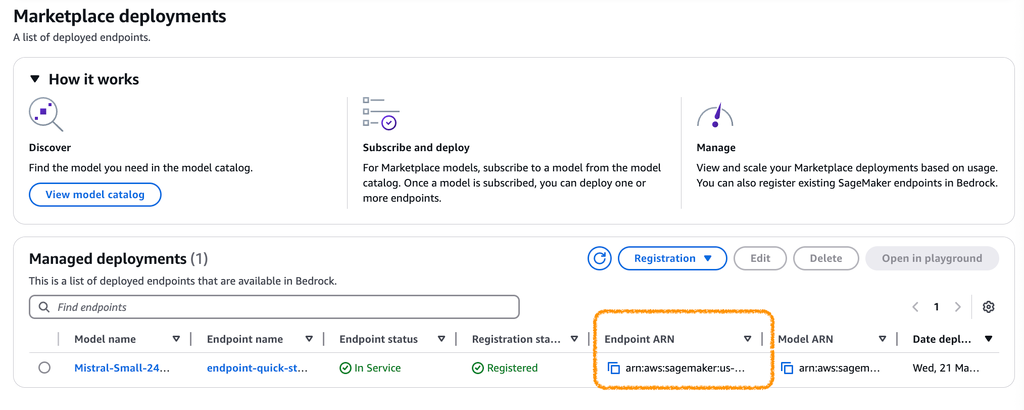
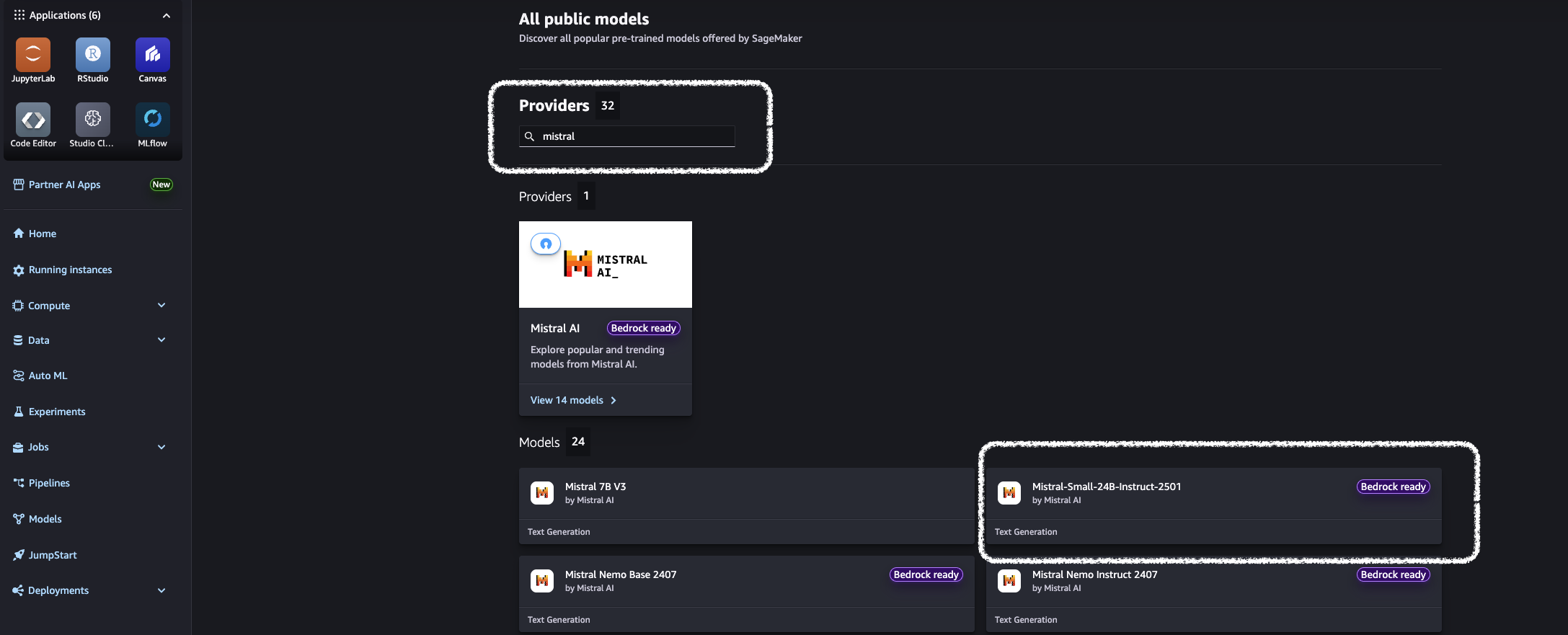
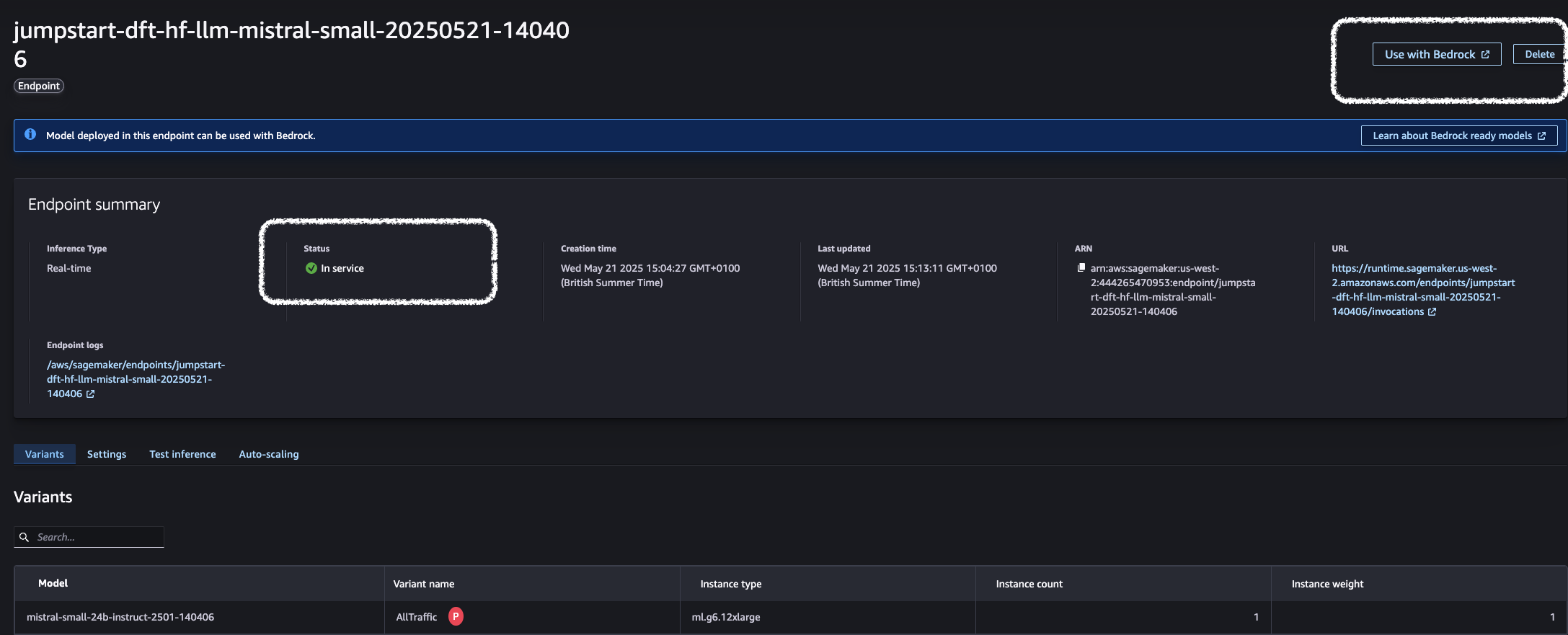
 Ying Hou, PhD, is a Sr. Specialist Solution Architect for Gen AI at AWS, where she collaborates with model providers to onboard the latest and most intelligent AI models onto AWS platforms. With deep expertise in Gen AI, ASR, computer vision, NLP, and time-series forecasting models, she works closely with customers to design and build cutting-edge ML and GenAI applications.
Ying Hou, PhD, is a Sr. Specialist Solution Architect for Gen AI at AWS, where she collaborates with model providers to onboard the latest and most intelligent AI models onto AWS platforms. With deep expertise in Gen AI, ASR, computer vision, NLP, and time-series forecasting models, she works closely with customers to design and build cutting-edge ML and GenAI applications. Siddhant Waghjale, is an Applied AI Engineer at Mistral AI, where he works on challenging customer use cases and applied science, helping customers achieve their goals with Mistral models. He’s passionate about building solutions that bridge AI capabilities with actual business applications, specifically in agentic workflows and code generation.
Siddhant Waghjale, is an Applied AI Engineer at Mistral AI, where he works on challenging customer use cases and applied science, helping customers achieve their goals with Mistral models. He’s passionate about building solutions that bridge AI capabilities with actual business applications, specifically in agentic workflows and code generation. Samuel Barry is an Applied AI Engineer at Mistral AI, where he helps organizations design, deploy, and scale cutting-edge AI systems. He partners with customers to deliver high-impact solutions across a range of use cases, including RAG, agentic workflows, fine-tuning, and model distillation. Alongside engineering efforts, he also contributes to applied research initiatives that inform and strengthen production use cases.
Samuel Barry is an Applied AI Engineer at Mistral AI, where he helps organizations design, deploy, and scale cutting-edge AI systems. He partners with customers to deliver high-impact solutions across a range of use cases, including RAG, agentic workflows, fine-tuning, and model distillation. Alongside engineering efforts, he also contributes to applied research initiatives that inform and strengthen production use cases. Preston Tuggle is a Sr. Specialist Solutions Architect with the Third-Party Model Provider team at AWS. He focuses on working with model providers across Amazon Bedrock and Amazon SageMaker, helping them accelerate their go-to-market strategies through technical scaling initiatives and customer engagement.
Preston Tuggle is a Sr. Specialist Solutions Architect with the Third-Party Model Provider team at AWS. He focuses on working with model providers across Amazon Bedrock and Amazon SageMaker, helping them accelerate their go-to-market strategies through technical scaling initiatives and customer engagement.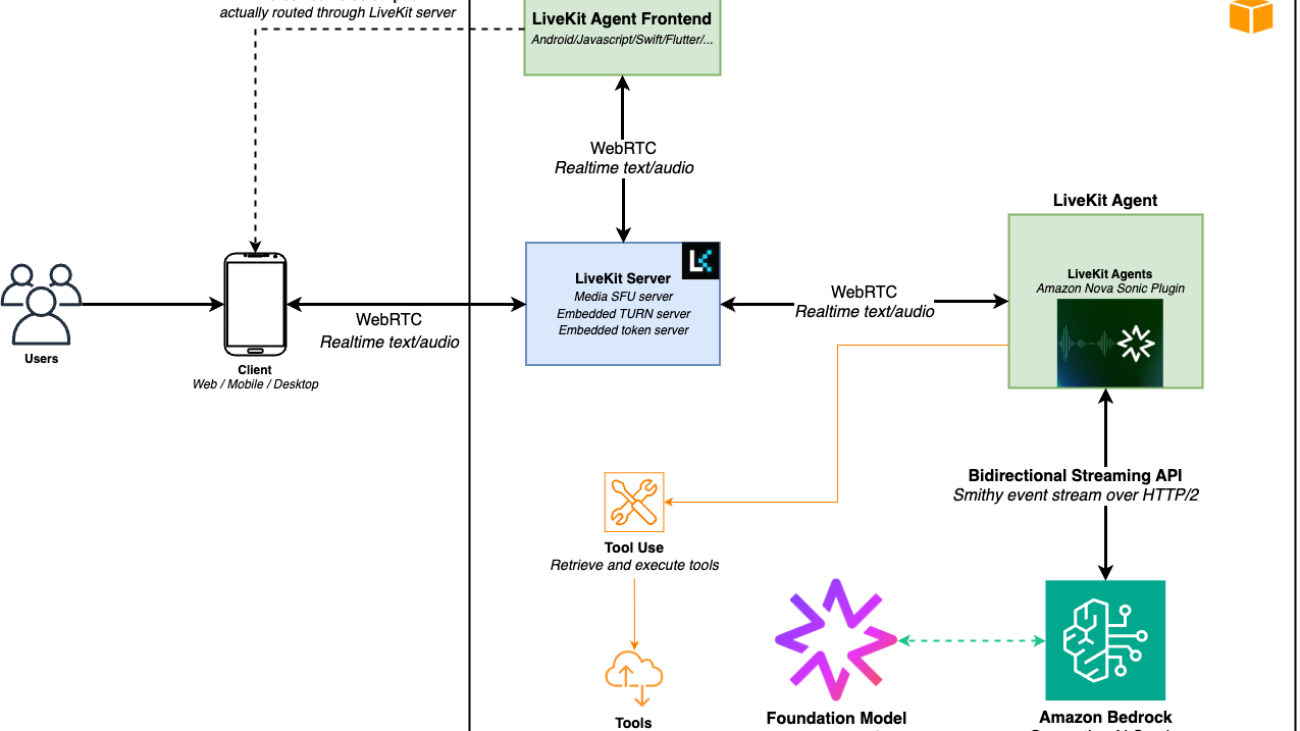








 Which game demands the fastest reflexes you’ve got?
Which game demands the fastest reflexes you’ve got? NVIDIA GeForce NOW (@NVIDIAGFN)
NVIDIA GeForce NOW (@NVIDIAGFN) 
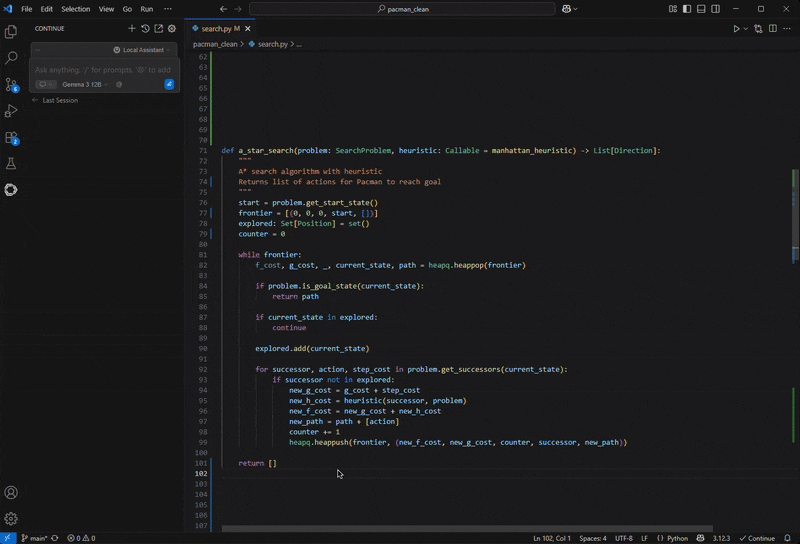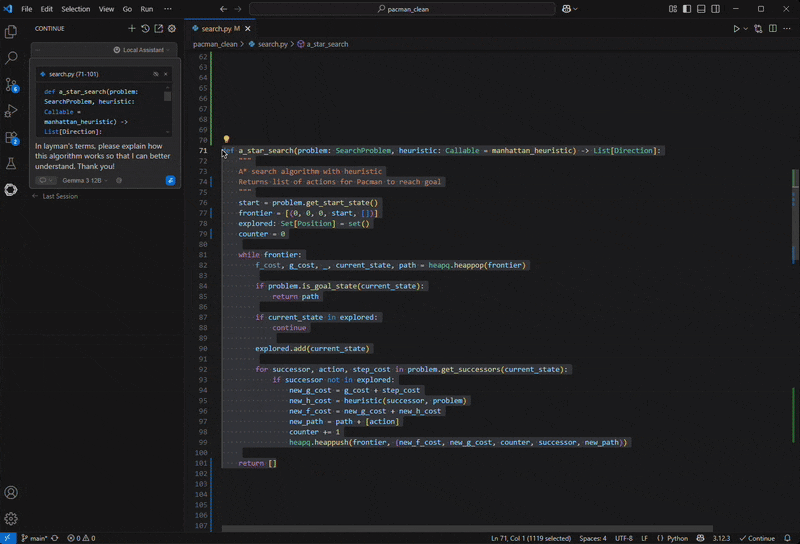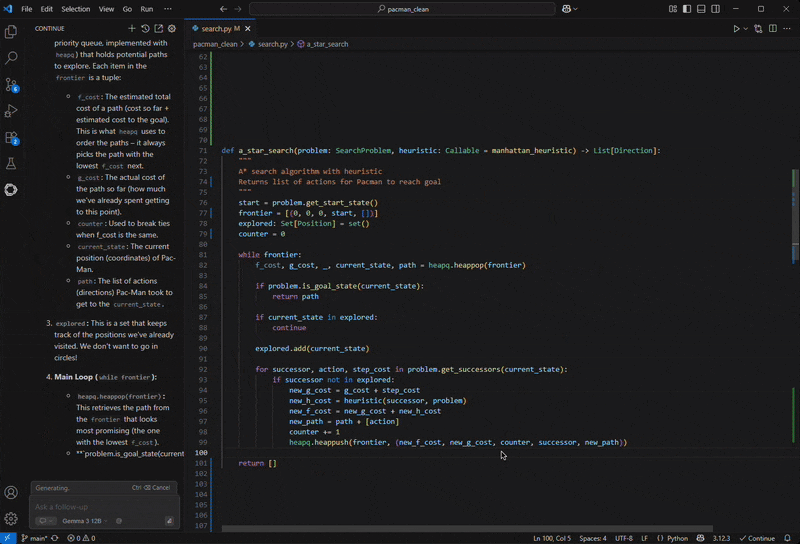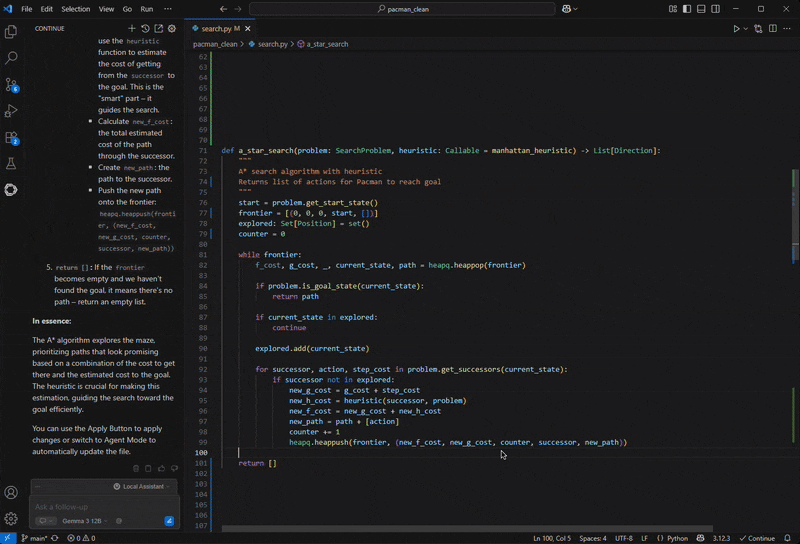

 David Brown is the Vice President of AWS Compute and Machine Learning (ML) Services. In this role he is responsible for building all AWS Compute and ML services, including Amazon EC2, Amazon Container Services, AWS Lambda, Amazon Bedrock and Amazon SageMaker. These services are used by all AWS customers but also underpin most of AWS’s internal Amazon applications. He also leads newer solutions, such as AWS Outposts, that bring AWS services into customers’ private data centers.
David Brown is the Vice President of AWS Compute and Machine Learning (ML) Services. In this role he is responsible for building all AWS Compute and ML services, including Amazon EC2, Amazon Container Services, AWS Lambda, Amazon Bedrock and Amazon SageMaker. These services are used by all AWS customers but also underpin most of AWS’s internal Amazon applications. He also leads newer solutions, such as AWS Outposts, that bring AWS services into customers’ private data centers.