According to an article by Cybersecurity Ventures, the damage caused by Ransomware (a type of malware that can block users from accessing their data unless they pay a ransom) increased by 57 times in 2021 as compared to 2015. Furthermore, it’s predicted to cost its victims $265 billion (USD) annually by 2031. At the time of writing, the financial toll from Ransomware attacks falls just above the 50th position in a list of countries ranked by their GDP.
Given the threat posed by malware, several techniques have been developed to detect and contain malware attacks. The two most common techniques used today are signature- and behavior-based detection.
Signature-based detection establishes a unique identifier about a known malicious object so that the object can be identified in the future. It may be a unique pattern of code attached to a file, or it may be the hash of a known malware code. If a known pattern identifier (signature) is discovered while scanning new objects, then the object is flagged as malicious. Signature-based detection is fast and requires low compute power. However, it struggles against polymorphic malware types, which continuously change their form to evade detection.
Behavior-based detection judges the suspicious objects based on their behavior. Artifacts that may be considered by anti-malware products are process interactions, DNS queries, and network connections from the object. This technique performs better at detecting polymorphic malware as compared to signature-based, but it does have some downsides. To assess if an object is malicious, it must run on the host and generate enough artifacts for the anti-malware product to detect it. This blind spot can let the malware infect the host and spread through the network.
Existing techniques are far from perfect. As a result, research continues with the aim to develop new alternative techniques that will improve our capabilities to combat against malware. One novel technique that has emerged in recent years is image-based malware detection. This technique proposes to train a deep-learning network with known malware binaries converted in greyscale images. In this post, we showcase how to perform Image-based Malware detection with Amazon Rekognition Custom Labels.
Solution overview
To train a multi-classification model and a malware-detection model, we first prepare the training and test datasets which contain different malware types such as flooder, adware, spyware, etc., as well as benign objects. We then convert the portable executables (PE) objects into greyscale images. Next, we train a model using the images with Amazon Rekognition.
Amazon Rekognition is a service that makes it simple to perform different types of visual analysis on your applications. Rekognition Image helps you build powerful applications to search, verify, and organize millions of images.
Amazon Rekognition Custom Labels builds off of Rekognition’s existing capabilities, which are already trained on tens of millions of images across many categories.
Amazon Rekognition Custom Labels is a fully-managed service that lets users analyze millions of images and utilize them to solve many different machine learning (ML) problems, including image classification, face detection, and content moderations. Behind the scenes, Amazon Rekognition is based on a deep learning technology. The service employs a convolution neural network (CNN), which is pre-trained on a large labeled dataset. By being exposed to such ground truth data, the algorithm can learn to recognize patterns in images from many different domains and can be used across many industry use-cases. Since AWS takes ownership of building and maintaining the model architecture and selecting an appropriate training method to the task at hand, users don’t need to spend time managing the infrastructure required for training tasks.
Solution architecture
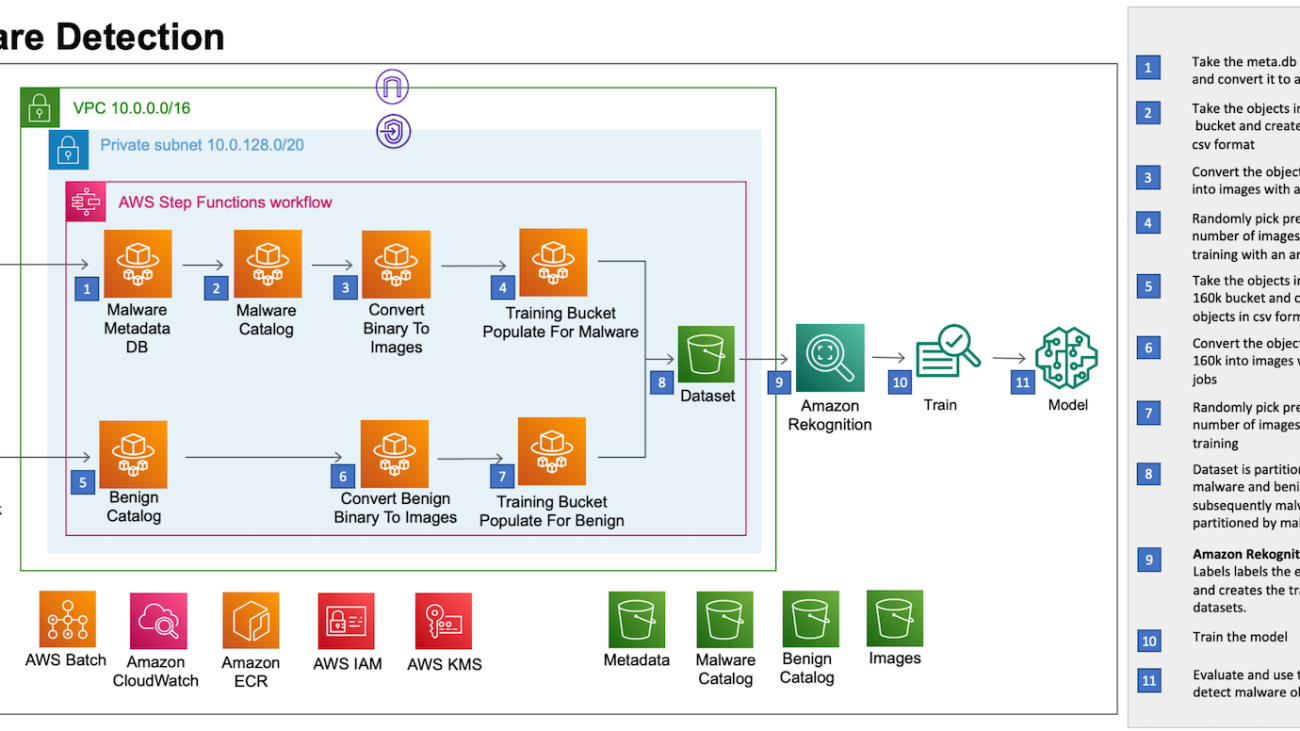
The following architecture diagram provides an overview of the solution.

The solution is built using AWS Batch, AWS Fargate, and Amazon Rekognition. AWS Batch lets you run hundreds of batch computing jobs on Fargate. Fargate is compatible with both Amazon Elastic Container Service (Amazon ECS) and Amazon Elastic Kubernetes Service (Amazon EKS). Amazon Rekognition custom labels lets you use AutoML for computer vision to train custom models to detect malware and classify various malware categories. AWS Step Functions are used to orchestrate data preprocessing.
For this solution, we create the preprocessing resources via AWS CloudFormation. The CloudFormation stack template and the source code for the AWS Batch, Fargate, and Step functions are available in a GitHub Repository.
Dataset
To train the model in this example, we used the following public datasets to extract the malicious and benign Portable Executable (PE):
- Sophos/ReversingLabs 20 Million malware detection dataset provides an extensive data set of malware objects from 11 different families. Since the dataset already exists on Amazon Simple Storage Service (Amazon S3), it’s relatively simple to embed it in the preprocessing flow.
- We source the benign objects through PE Malware Machine Learning Dataset, created and owned by Michael Lester. If you plan to use the same dataset, then first you will manually download the dataset and upload the binaries into an S3 bucket (i.e., benign-160k) which feeds the preprocessing flow.
We encourage you to read carefully through the datasets documentation (Sophos/Reversing Labs README, PE Malware Machine Learning Dataset) to safely handle the malware objects. Based on your preference, you can also use other datasets as long as they provide malware and benign objects in the binary format.
Next, we’ll walk you through the following steps of the solution:
- Preprocess objects and convert to images
- Deploy preprocessing resources with CloudFormation
- Choose the model
- Train the model
- Evaluate the model
- Cost and performance
Preprocess objects and convert to images
We use Step Functions to orchestrate the object preprocessing workflow which includes the following steps:
- Take the meta.db sqllite database from sorel-20m S3 bucket and convert it to a .csv file. This helps us load the .csv file in a Fargate container and refer to the metadata while processing the malware objects.
- Take the objects from the sorel-20m S3 bucket and create a list of objects in the csv format. By performing this step, we’re creating a series of .csv files which can be processed in parallel, thereby reducing the time taken for the preprocessing.
- Convert the objects from the sorel-20m S3 bucket into images with an array of jobs. AWS Batch array jobs share common parameters for converting the malware objects into images. They run as a collection of image conversion jobs that are distributed across multiple hosts, and run concurrently.
- Pick a predetermined number of images for the model training with an array of jobs corresponding to the categories of malware.
- Similar to Step 2, we take the benign objects from the benign-160k S3 bucket and create a list of objects in csv format.
- Similar to Step 3, we convert the objects from the benign-160k S3 bucket into images with an array of jobs.
- Due to the Amazon Rekognition default quota for custom labels training (250K images), pick a predetermined number of benign images for the model training.
- As shown in the following image, the images are stored in an S3 bucket partitioned first by malware and benign folders, and then subsequently the malware is partitioned by malware types.


Deploy the preprocessing resources with CloudFormation
Prerequisites
The following prerequisites are required before continuing:
- An AWS Account
- AWS Command Line Interface (AWS CLI)
- Python
- Docker
- AWS SDK for Python
- Amazon S3 Buckets that contain benign objects and their metadata database
- Amazon S3 Buckets that contain artifacts
Resource deployment
The CloudFormation stack will create the following resources:
- An Amazon Virtual Private Cloud (Amazon VPC)
- Public and Private Subnets
- Internet Gateway
- Nat Gateway
- AWS Batch Compute Environment
- AWS Batch Queues
- AWS Batch Job Definitions
- AWS Step Functions
- Amazon S3 Buckets
- AWS Key Management Service (AWS KMS) key
- AWS Identity and Access Management (IAM) roles and policies
- Amazon CloudWatch Logs Group
Parameters
- STACK_NAME – CloudFormation stack name
- AWS_REGION – AWS region where the solution will be deployed
- AWS_PROFILE – Named profile that will apply to the AWS CLI command
- ARTEFACT_S3_BUCKET – S3 bucket where the infrastructure code will be stored. (The bucket must be created in the same region where the solution lives).
- AWS_ACCOUNT – AWS Account ID.
Use the following commands to deploy the resources
Make sure the docker agent is running on the machine. The deployments are done using bash scripts, and in this case we use the following command:
bash malware_detection_deployment_scripts/deploy.sh -s '<STACK_NAME>' -b 'malware-
detection-<ACCOUNT_ID>-artifacts' -p <AWS_PROFILE> -r "<AWS_REGION>" -a
<ACCOUNT_ID>This builds and deploys the local artifacts that the CloudFormation template (e.g., cloudformation.yaml) is referencing.
Train the model
Since Amazon Rekognition takes care of model training for you, computer vision or highly specialized ML knowledge isn’t required. However, you will need to provide Amazon Rekognition with a bucket filled with appropriately labeled input images.
In this post, we’ll train two independent image classification models via the custom labels feature:
- Malware detection model (binary classification) – identify if the given object is malicious or benign
- Malware classification model (multi-class classification) – identify the malware family for a given malicious object
Model training walkthrough
The steps listed in the following walkthrough apply to both models. Therefore, you will need to go through the steps two times in order to train both models.
- Sign in to the AWS Management Console and open the Amazon Rekognition console.
- In the left pane, choose Use Custom Labels. The Amazon Rekognition Custom Labels landing page is shown.
- From the Amazon Rekognition Custom Labels landing page, choose Get started.
- In the left pane, Choose Projects.
- Choose Create Project.
- In Project name, enter a name for your project.
- Choose Create project to create your project.
- In the Projects page, choose the project to which you want to add a dataset. The details page for your project is displayed.
- Choose Create dataset. The Create dataset page is shown.
- In Starting configuration, choose Start with a single dataset to let Amazon Rekognition split the dataset to training and test. Note that you might end up with different test samples in each model training iteration, resulting in slightly different results and evaluation metrics.
- Choose Import images from Amazon S3 bucket.
- In S3 URI, enter the S3 bucket location and folder path. The same S3 bucket provided from the preprocessing step is used to create both datasets: Malware detection and Malware classification. The Malware detection dataset points to the root (i.e.,
s3://malware-detection-training-{account-id}-{region}/) of the S3 bucket, while the Malware classification dataset points to the malware folder (i.e.,s3://malware-detection-training-{account-id}-{region}/malware) of the S3 bucket.
- Choose Automatically attach labels to images based on the folder.
- Choose Create Datasets. The datasets page for your project opens.
- On the Train model page, choose Train model. The Amazon Resource Name (ARN) for your project should be in the Choose project edit box. If not, then enter the ARN for your project.
- In the Do you want to train your model? dialog box, choose Train model.
- After training completes, choose the model’s name. Training is finished when the model status is TRAINING_COMPLETED.
- In the Models section, choose the Use model tab to start using the model.
For more details, check the Amazon Rekognition custom labels Getting started guide.
Evaluate the model
When the training models are complete, you can access the evaluation metrics by selecting Check metrics on the model page. Amazon Rekognition provides you with the following metrics: F1 score, average precision, and overall recall, which are commonly used to evaluate the performance of classification models. The latter are averaged metrics over the number of labels.
In the Per label performance section, you can find the values of these metrics per label. Additionally, to get the values for True Positive, False Positive, and False negative, select the View test results.
Malware detection model metrics
On the balanced dataset of 199,750 images with two labels (benign and malware), we received the following results:
- F1 score – 0.980
- Average precision – 0.980
- Overall recall – 0.980
Malware classification model metrics
On the balanced dataset of 130,609 images with 11 labels (11 malware families), we received the following results:
- F1 score – 0.921
- Average precision – 0.938
- Overall recall – 0.906
To assess whether the model is performing well, we recommend comparing its performance with other industry benchmarks which have been trained on the same (or at least similar) dataset. Unfortunately, at the time of writing of this post, there are no comparative bodies of research which solve this problem using the same technique and the same datasets. However, within the data science community, a model with an F1 score above 0.9 is considered to perform very well.
Cost and performance
Due to the serverless nature of the resources, the overall cost is influenced by the amount of time that each service is used. On the other hand, performance is impacted by the amount of data being processed and the training dataset size feed to Amazon Rekognition. For our cost and performance estimate exercise, we consider the following scenario:
- 20 million objects are cataloged and processed from the sorel dataset.
- 160,000 objects are cataloged and processed from the PE Malware Machine Learning Dataset.
- Approximately 240,000 objects are written to the training S3 bucket: 160,000 malware objects and 80,000 benign objects.
Based on this scenario, the average cost to preprocess and deploy the models is $510.99 USD. You will be charged additionally $4 USD/h for every hour that you use the model. You may find the detailed cost breakdown in the estimate generated via the AWS Pricing Calculator.
Performance-wise, these are the results from our measurement:
- ~2 h for the preprocessing flow to complete
- ~40 h for the malware detecting model training to complete
- ~40 h for the malware classification model training to complete
Clean-up
To avoid incurring future charges, stop and delete the Amazon Rekognition models, and delete the preprocessing resources via the destroy.sh script. The following parameters are required to run the script successfully:
- STACK_NAME – The CloudFormation stack name
- AWS_REGION – The Region where the solution is deployed
- AWS_PROFILE – The named profile that applies to the AWS CLI command
Use the following commands to run the ./malware_detection_deployment_scripts/destroy.sh script:
bash malware_detection_deployment_scripts/destroy.sh -s <STACK_NAME> -p
<AWS_PROFILE> -r <AWS_REGION>Conclusion
In this post, we demonstrated how to perform malware detection and classification using Amazon Rekognition. The solutions follow a serverless pattern, leveraging managed services for data preprocessing, orchestration, and model deployment. We hope that this post helps you in your ongoing efforts to combat malware.
In a future post we’ll show a practical use case of malware detection by consuming the models deployed in this post.
About the authors
 Edvin Hallvaxhiu is a Senior Global Security Architect with AWS Professional Services and is passionate about cybersecurity and automation. He helps customers build secure and compliant solutions in the cloud. Outside work, he likes traveling and sports.
Edvin Hallvaxhiu is a Senior Global Security Architect with AWS Professional Services and is passionate about cybersecurity and automation. He helps customers build secure and compliant solutions in the cloud. Outside work, he likes traveling and sports.
 Rahul Shaurya is a Principal Data Architect with AWS Professional Services. He helps and works closely with customers building data platforms and analytical applications on AWS. Outside of work, Rahul loves taking long walks with his dog Barney.
Rahul Shaurya is a Principal Data Architect with AWS Professional Services. He helps and works closely with customers building data platforms and analytical applications on AWS. Outside of work, Rahul loves taking long walks with his dog Barney.
 Bruno Dhefto is a Global Security Architect with AWS Professional Services. He is focused on helping customers building Secure and Reliable architectures in AWS. Outside of work, he is interested in the latest technology updates and traveling.
Bruno Dhefto is a Global Security Architect with AWS Professional Services. He is focused on helping customers building Secure and Reliable architectures in AWS. Outside of work, he is interested in the latest technology updates and traveling.
 Nadim Majed is a data architect within AWS professional services. He works side by side with customers building their data platforms on AWS. Outside work, Nadim plays table tennis, and loves watching football/soccer.
Nadim Majed is a data architect within AWS professional services. He works side by side with customers building their data platforms on AWS. Outside work, Nadim plays table tennis, and loves watching football/soccer.













































 Vadim Omeltchenko is a Sr. AI/ML Solutions Architect who is passionate about helping AWS customers innovate in the cloud. His prior IT experience was predominantly on the ground.
Vadim Omeltchenko is a Sr. AI/ML Solutions Architect who is passionate about helping AWS customers innovate in the cloud. His prior IT experience was predominantly on the ground.


 Dr. Taha Kass-Hout is Vice President, Machine Learning, and Chief Medical Officer at Amazon Web Services, and leads our Health AI strategy and efforts, including Amazon Comprehend Medical and Amazon HealthLake. He works with teams at Amazon responsible for developing the science, technology, and scale for COVID-19 lab testing, including Amazon’s first FDA authorization for testing our associates—now offered to the public for at-home testing. A physician and bioinformatician, Taha served two terms under President Obama, including the first Chief Health Informatics officer at the FDA. During this time as a public servant, he pioneered the use of emerging technologies and the cloud (the CDC’s electronic disease surveillance), and established widely accessible global data sharing platforms: the openFDA, which enabled researchers and the public to search and analyze adverse event data, and precisionFDA (part of the Presidential Precision Medicine initiative).
Dr. Taha Kass-Hout is Vice President, Machine Learning, and Chief Medical Officer at Amazon Web Services, and leads our Health AI strategy and efforts, including Amazon Comprehend Medical and Amazon HealthLake. He works with teams at Amazon responsible for developing the science, technology, and scale for COVID-19 lab testing, including Amazon’s first FDA authorization for testing our associates—now offered to the public for at-home testing. A physician and bioinformatician, Taha served two terms under President Obama, including the first Chief Health Informatics officer at the FDA. During this time as a public servant, he pioneered the use of emerging technologies and the cloud (the CDC’s electronic disease surveillance), and established widely accessible global data sharing platforms: the openFDA, which enabled researchers and the public to search and analyze adverse event data, and precisionFDA (part of the Presidential Precision Medicine initiative).




















 Compare data processing job results
Compare data processing job results