Amazon SageMaker is a fully-managed service that provides every developer and data scientist with the ability to quickly build, train, and deploy machine learning (ML) models at scale. ML is realized in inference. SageMaker offers four Inference options:
These four options can be broadly classified into Online and Batch inference options. In Online Inference, requests are expected to be processed as they arrive, and the consuming application expects a response after each request is processed. This can either happen synchronously (real-time Inference, serverless) or asynchronously (asynchronous inference). In a synchronous pattern, the consuming application is blocked and can’t proceed until it receives a response. These workloads tend to be real-time applications, such as online credit card fraud detection, where responses are expected in the order of milliseconds to seconds and request payloads are small (a few MB). In the asynchronous pattern, the application experience isn’t blocked (for example, submitting an insurance claim via a mobile app), and usually requires larger payload sizes and/or longer processing times. In Offline inference, an aggregation (batch) of inference requests are processed together, and responses are provided only after the entire batch has been processed. Usually, these workloads aren’t latency sensitive, involve large volumes (multiple GBs) of data, and are scheduled at a regular cadence (for example, run object detection on security camera footage at the end of the day or process payroll data at the end of the month).
At the bare bones, SageMaker Real-Time Inference consists of a model(s), the framework/container with which you’re working, and the infrastructure/instances that are backing your deployed endpoint. In this post, we’ll explore how you can create and invoke a Single Model Endpoint.
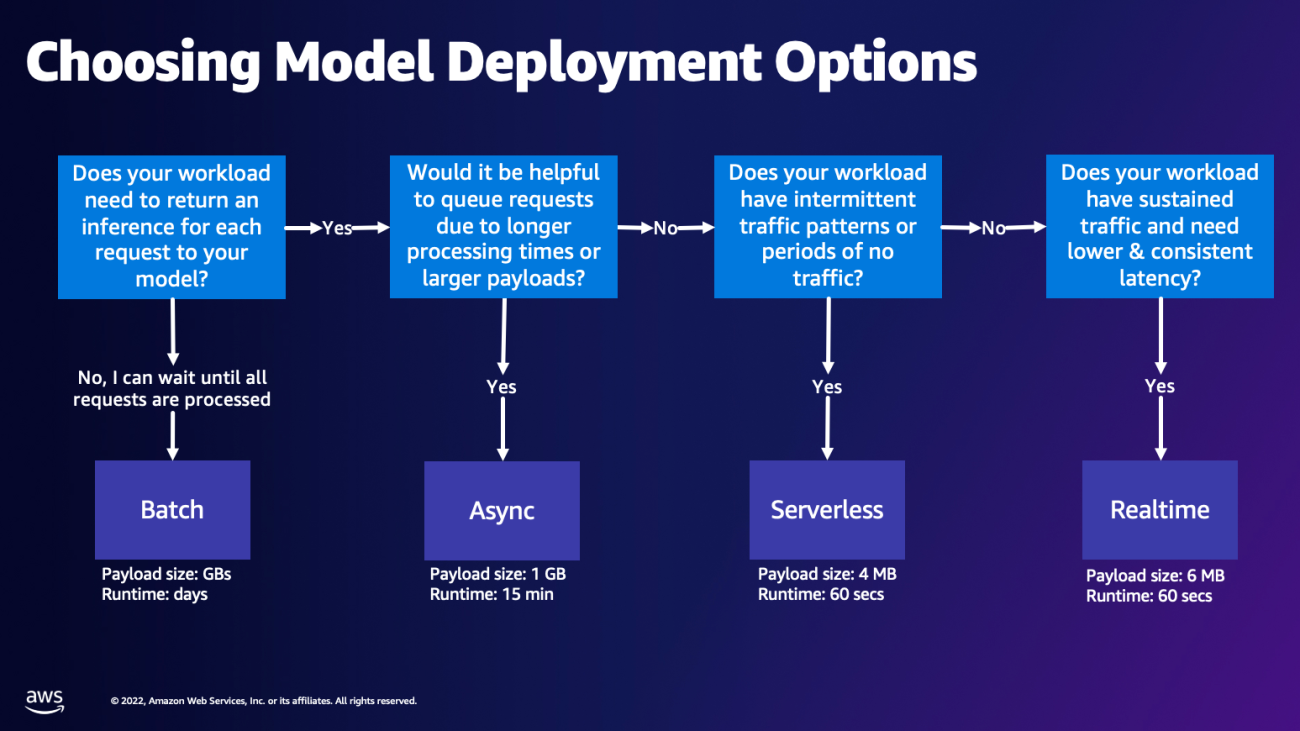
Choosing model deployment option
Choosing the right inference type can be difficult, and the following simple guide can help you. It’s not a strict flow chart, so if you find that another option works better for you, then feel free to use those. In particular, Real-Time Inference is a great option for hosting your models when you have low and consistent latency (in the order of milliseconds or seconds) and throughput sensitive workloads. You can control the instance type and count behind your endpoint while also configuring AutoScaling policy to handle traffic. There are two other SageMaker Inference options that you can also use to create an endpoint. Asynchronous Inference is when you have large payload sizes and near real-time latency bandwidth. This is a good option, especially for NLP and Computer Vision models that have longer preprocessing times. Serverless Inference is a great option when you have intermittent traffic and don’t want to manage infrastructure scaling. The recipe for creating an endpoint remains the same regardless of the Inference type that you choose. In this post, we’ll focus on creating a real-time instance-based endpoint, but you can easily adapt it to either of the other Inference Options based on your use-case. Lastly, Batch inference takes place offline, so you can provide a set of data that you want to get inference from and we’ll run it. This is similarly instance-based, so you can select the optimal instance for your workload. As there is no endpoint up and running, you only pay for the duration of the job. It is good for processing gigabytes of data and the job duration can be days. There are built-in features to make working with structured data easier and optimizations to automatically distribute structured data. Some example use cases are propensity modeling, predictive maintenance, and churn prediction. All of these can take place offline in bulk because it doesn’t have to react to a specific event.
Hosting a model on SageMaker Endpoints
At the crux, SageMaker Real-Time Endpoints consists of a model and the infrastructure with which you choose to back the Endpoint. SageMaker uses containers to host models, which means that you need a container that properly sets up the environment for the framework that you use for each model that you provide. For example, if you’re working with a Sklearn model, you must pass in your model scripts/data within a container that properly sets up Sklearn. Luckily, SageMaker provides managed images for popular frameworks, such as TensorFlow, PyTorch, Sklearn, and HuggingFace. You can retrieve and utilize these images using the high-level SageMaker Python SDK and inject your model scripts and data into these containers. In the case that SageMaker doesn’t have a supported container, you can also Build Your Own Container and push your own custom image, installing the dependencies that are necessary for your model.
SageMaker supports both trained and pre-trained models. In the previous paragraph when we’re talking about model scripts/data, we’re referencing this matter. You can either mount a script on your container, or if you have a pre-trained model artifact (for example, `model.joblib` for SKLearn), then you can provide this along with your image to SageMaker. To understand SageMaker Inference, there are three main entities that you’ll create in the process of Endpoint creation:
- SageMaker Model Entity – Here you can pass in your trained model data/model script and your image that you’re working with, whether it’s owned by AWS or built by you.
- Endpoint configuration creation – Here you define your infrastructure, meaning that you select the instance type, count, etc.
- Endpoint creation – This is the REST Endpoint that hosts your model that you’re invoking to get a response. Let’s look at how you can utilize a managed SageMaker Image and your own custom-built image to deploy an endpoint.
Real-time endpoint requirements
- Before creating an Endpoint, you must understand what type of Model you want to host. If it’s a Framework model, such as TensorFlow, PyTorch, or MXNet, then you can utilize one of the prebuilt Framework images.
If it’s a custom model, or you would like full flexibility in creating the container that SageMaker will run for inference, then you can build your own container.
SageMaker Endpoints are made up of a SageMaker Model and Endpoint Configuration.
If you’re using Boto3, then you would create both objects. Otherwise, if you’re utilizing the SageMaker Python SDK, then the Endpoint Configuration is created on your behalf when you use the .deploy(..) function.
SageMaker entities:
- SageMaker Model:
- Contains the details of the inference image, location of the model artifacts in Amazon Simple Storage Service (Amazon S3), network configuration, and AWS Identity and Access Management (IAM) role to be used by the Endpoint.
- SageMaker requires your model artifacts to be compressed in a
.tar.gzfile. SageMaker automatically extracts this.tar.gzfile into the/opt/ml/model/directory in your container. If you’re utilizing one of the framework containers, such as TensorFlow, PyTorch, or MXNet, then the container expects your TAR structure to be as follows:- TensorFlow
- PyTorch
- MXNet
- Sklearn
- TensorFlow
- When utilizing a Framework image, we can provide a custom entry-point script, where we can implement our own pre and post processing. In our case, the inference script is packaged in the model.tar.gz under the /code directory.
- SageMaker requires your model artifacts to be compressed in a
- Contains the details of the inference image, location of the model artifacts in Amazon Simple Storage Service (Amazon S3), network configuration, and AWS Identity and Access Management (IAM) role to be used by the Endpoint.
-
-
- Endpoint Configuration
- Contains the infrastructure information required to deploy the SageMaker Model to the Endpoint.
- For example, the SageMaker Model we created is specified here as well as the Instance Type and Initial Instance count.
-
Frameworks and BYOC
-
- Retrieving SageMaker images
- This portion isn’t always necessary and abstracted out by the SageMaker Python SDK via estimators. However, if you would like to be able to retrieve a SageMaker managed image to extend on it, then you can get the images that are available via the SDK. The following is an example of retreiving a TF 2.2 image for inference.
- Frameworks
- In the case that you want to deploy a Framework Model, such as TensorFlow, PyTorch, or MXNet, then all you need is the model artifacts.
- See the documentation for deploying models directly from model artifacts for TensorFlow, PyTorch, or MXNet.
- Retrieving SageMaker images
-
- Choosing between 1P and BYOC
- The SageMaker SDK also abstracts handling the image out, as you saw in the previous Frameworks section. It has ready-made estimators for Sklearn, TensorFlow, and PyTorch that automatically select the image for you based off of the version that you’ve selected. Then you can pass in a training/inference script through Script Mode into these estimators.
- Not all packages and images are supported by SageMaker, and in this case you must bring your own container (BYOC). This means building a Dockerfile that will setup the proper environment for your model serving. An example of this is the Spacy NLP module, and there are no managed SageMaker containers for this framework. Therefore, you must provide a Dockerfile that installs Spacy. Within the container you also mount your model inference scripts. Let’s quickly discuss the components that you provide in a Bring Your Own Container format, as these stay consistent for most examples.
- “nginx.conf“ is the configuration file for the nginx front-end. You won’t have to edit this file, unless you would like to tune these portions.
- “predictor.py“ is the program that actually implements the Flask web server and model code for your application. You can have further Python files or functions in your container that you can call in this file.
- “serve“ is the program started when the container is started for hosting. It simply launches the gunicorn server, which runs multiple instances of the Flask app defined in predictor.py. Like nginx.conf, you don’t have to edit this file unless there’s further tuning that you would like to perform.
- “train“ is the program that is invoked when the container is run for training. You’ll modify this program to implement your training algorithm. If you’re bringing a pre-trained model or framework like Spacy, then you don’t need this file.
- “wsgi.py“ is a small wrapper used to invoke the Flask app. You should be able to take this file as-is, unless you’ve changed the name of your predictor.py file. In that case, make sure that maps properly here.
- Choosing between 1P and BYOC
-
- Custom inference script
- SageMaker Framework containers give you the flexibility to handle pre/post processing of the request and model loading using a custom entry point script/inference.py.
- See the documentation for creating a custom inference.py script for TensorFlow, PyTorch and MXNet.
- Custom container
- Custom containers allow you full autonomy in using your own inference code to host your model. Your container is required to comply with certain API contracts. For example, it must respond to /invocations and /ping on port 8080. This Scikit Bring Your Own Container example showcases a common hosting pattern of NGINXGunicornFlask.
Different ways that you can interact with SageMaker Endpoints
There are many options for using SageMaker programmatically so that you can call your deployed models to get inference. The AWS Command Line Interface (AWS CLI), REST APIs, AWS CloudFormation, AWS Cloud Development Kit (AWS CDK), and AWS SDKs are common tools offered by AWS and widely supported by other AWS services. For SageMaker, we also have a SageMaker Python SDK. Now, let’s compare the different options to create, invoke, and manage SageMaker Endpoints.
In addition to SageMaker CLI, there are two ways programmatically that you can interact with Endpoints in SageMaker through the SDKs. Let’s look at some differences between SageMaker Python SDK and Boto3 Python SDK:
- High-level SageMaker “Python” SDK – This SDK is an open-source library that provides higher level abstraction specifically meant for calling SageMaker APIs programmatically using Python. The Good part of this SDK is that it’s very easy to call sagemaker APIs, lots of heavy lifting is done already like calling the APIs synchronously/async mode (helps to avoid polling), simpler request/response schema, much less code, and much simpler code. SageMaker Python SDK provides several high-level abstractions for working with SageMaker. The package is meant to simplify different ML processes on SageMaker.
- Low-level AWS SDK (Boto3 SDK) – This SDK works at the lower level by allowing the user to choose from the supported programming languages and call any AWS services programmatically. This isn’t just specific to SageMaker but can be used in general for all AWS services. The low-level AWS SDKs are available in various programming languages, such as .NET, Python, Java, Node.js, etc. One of the popular SDKs used is boto3 python SDK, which is popular in the data scientist community for ML. The good part of this SDK is that it’s very lightweight and available by default installed on AWS Lambda Runtime. Furthermore, you can use this SDK to interact with any AWS service outside of SageMaker.
Both of these SDKs can be utilized for the same tasks, but in some cases it’s more intuitive to use one more than the other. SageMaker Python SDK is recommended for easy testing while AWS SDK/Boto3 is recommended for production use cases for better control on performance. For example, SageMaker as a service provides pre-built and maintained images for popular frameworks, such as Sklearn, PyTorch, and TensorFlow. It can be particularly useful to use SageMaker SDK to retrieve deep learning images, train models using Estimators, and easily deploy the model using a simple API call. An example to showcase this in action can be found here.
On the other hand, sometimes you have pre-trained models or different frameworks that you may be using. This requires a greater deal of customization and the SageMaker SDK doesn’t always offer that. We have three important steps and corresponding boto3 API calls that we need to execute to deploy an endpoint: Model Creation, Endpoint Configuration Creation, and Endpoint Creation. The first two entities were abstracted out with the SageMaker SDK with our supported frameworks, but we see those details with the Boto3 SDK. An extensive example to showcase the steps involved in using a Boto3 SDK to create and manage an endpoint can be found here.
Considerations of SageMaker hosting
SageMaker Real-Time Inference has two main optimizations that you can consider: 1/ Performance optimization, and 2/ Cost optimization. Let’s first look at performance optimization, as when we’re dealing with latency sensitive workloads, every millisecond is crucial. There are different knobs that you can tune to optimize your latency and throughput. At the instance level, you can use Inference Recommender, our built-in load testing tool, to help you select the right instance type and count for your workload. Utilizing the proper combination of compute will help you with both performance and cost. You can also tune at the container and framework level.
Questions to ask yourself include:
- What framework are you using?
- Are there any environment variables that you can tune within your container?
An example of this is maximizing TensorFlow performance with SageMaker containers. Another example of container level optimizations is utilizing gRPC rather than REST behind your endpoint. Lastly, you can also optimize at the script level. Is your inference code taking extra time at certain blocks? Timing each and every line of your script will help you capture any bottlenecks within your code.
There are three ways to look at improving the utilization of your Real Time endpoint:
- Multi-model Endpoints (MME)
- You can host thousands of models behind a single endpoint. This is perfect for use cases where you don’t need a dedicated endpoint for each one of your models. MME works best when the models are similarly sized and latency and belong to the same ML framework. These can be typically used when you don’t need to call the same model at all times. You can dynamically load the respective model onto the SageMaker Endpoint to serve your request. An example that showcases MME in action can be found here. If you want to learn more about the different caveats and best practices for hosting models on MME, then refer to the post here.
- Multi-Container Endpoints (MCE)
- Instead of utilizing multiple endpoints to host multiple containers, you can look at hosting up to 15 containers on a single endpoint. Each one of these containers can be invoked directly. Therefore, you can look at hosting disparate models of different frameworks all on a single endpoint. This option is best when containers exhibit similar usage and performance characteristics. An example that showcases MCE can be found here. If you want to learn more about the different caveats and best practices for hosting models on MCE, then refer to the post here.
- Serial Inference Pipeline (SIP)
- If you have a pipeline of steps in your inference logic, then you might utilize Serial Inference Pipeline (SIP). SIP lets you chain 2-15 containers together behind a single endpoint. SIP works well when you have preprocessing and post-processing steps. If you want to learn more about the design patterns for serial inference pipelines, then refer to the post here.
The second main optimization to keep in mind is cost. Real-Time Inference is one of three options within creating SageMaker Endpoints. SageMaker Endpoints are running at all times unless deleted. Therefore, you must look at improving the utilization of the endpoint which in turn provides a cost benefit.
SageMaker also offers Savings Plans. Savings Plans can reduce your costs by up to 64%. This is a 1 or 3-year term commitment to a consistent amount of usage ($/hour). See this link for more information. And see this link for best to optimize costs for Inference on Amazon SageMaker.
Conclusion
In this post, we showed you some of the best practices to choose between different model hosting options on SageMaker. We discussed the SageMaker Endpoint requirements, and also contrasted Framework and BYOC requirements and functionality. Furthermore, we talked about the different ways that you can leverage Real-Time Endpoints to host your ML models in production. in a cost-effective way, and have high performance.
See the corresponding GitHub repository and try out the examples.
About the authors
 Raghu Ramesha is an ML Solutions Architect with the Amazon SageMaker Service team. He focuses on helping customers build, deploy, and migrate ML production workloads to SageMaker at scale. He specializes in machine learning, AI, and computer vision domains, and holds a master’s degree in Computer Science from UT Dallas. In his free time, he enjoys traveling and photography.
Raghu Ramesha is an ML Solutions Architect with the Amazon SageMaker Service team. He focuses on helping customers build, deploy, and migrate ML production workloads to SageMaker at scale. He specializes in machine learning, AI, and computer vision domains, and holds a master’s degree in Computer Science from UT Dallas. In his free time, he enjoys traveling and photography.
 Ram Vegiraju is a ML Architect with the SageMaker Service team. He focuses on helping customers build and optimize their AI/ML solutions on Amazon SageMaker. In his spare time, he loves traveling and writing.
Ram Vegiraju is a ML Architect with the SageMaker Service team. He focuses on helping customers build and optimize their AI/ML solutions on Amazon SageMaker. In his spare time, he loves traveling and writing.
 Marc Karp is a ML Architect with the SageMaker Service team. He focuses on helping customers design, deploy and manage ML workloads at scale. In his spare time, he enjoys traveling and exploring new places.
Marc Karp is a ML Architect with the SageMaker Service team. He focuses on helping customers design, deploy and manage ML workloads at scale. In his spare time, he enjoys traveling and exploring new places.
 Dhawal Patel is a Principal Machine Learning Architect at AWS. He has worked with organizations ranging from large enterprises to mid-sized startups on problems related to distributed computing and artificial intelligence. He focuses on deep learning, including NLP and computer vision domains. He helps customers achieve high-performance model inference on Amazon SageMaker.
Dhawal Patel is a Principal Machine Learning Architect at AWS. He has worked with organizations ranging from large enterprises to mid-sized startups on problems related to distributed computing and artificial intelligence. He focuses on deep learning, including NLP and computer vision domains. He helps customers achieve high-performance model inference on Amazon SageMaker.
 Saurabh Trikande is a Senior Product Manager for Amazon SageMaker Inference. He is passionate about working with customers and is motivated by the goal of democratizing machine learning. He focuses on core challenges related to deploying complex ML applications, multi-tenant ML models, cost optimizations, and making deployment of deep learning models more accessible. In his spare time, Saurabh enjoys hiking, learning about innovative technologies, following TechCrunch, and spending time with his family.
Saurabh Trikande is a Senior Product Manager for Amazon SageMaker Inference. He is passionate about working with customers and is motivated by the goal of democratizing machine learning. He focuses on core challenges related to deploying complex ML applications, multi-tenant ML models, cost optimizations, and making deployment of deep learning models more accessible. In his spare time, Saurabh enjoys hiking, learning about innovative technologies, following TechCrunch, and spending time with his family.
































 Rupinder Grewal is a Sr Ai/ML Specialist Solutions Architect with AWS. He currently focuses on serving of models and MLOps on SageMaker. Prior to this role he has worked as Machine Learning Engineer building and hosting models. Outside of work he enjoys playing tennis and biking on mountain trails.
Rupinder Grewal is a Sr Ai/ML Specialist Solutions Architect with AWS. He currently focuses on serving of models and MLOps on SageMaker. Prior to this role he has worked as Machine Learning Engineer building and hosting models. Outside of work he enjoys playing tennis and biking on mountain trails. Frank Liu is a Software Engineer for AWS Deep Learning. He focuses on building innovative deep learning tools for software engineers and scientists. In his spare time, he enjoys hiking with friends and family.
Frank Liu is a Software Engineer for AWS Deep Learning. He focuses on building innovative deep learning tools for software engineers and scientists. In his spare time, he enjoys hiking with friends and family.


 Qingwei Li is a Machine Learning Specialist at Amazon Web Services. He received his Ph.D. in Operations Research after he broke his advisor’s research grant account and failed to deliver the Nobel Prize he promised. Currently he helps customers in the financial service and insurance industry build machine learning solutions on AWS. In his spare time, he likes reading and teaching.
Qingwei Li is a Machine Learning Specialist at Amazon Web Services. He received his Ph.D. in Operations Research after he broke his advisor’s research grant account and failed to deliver the Nobel Prize he promised. Currently he helps customers in the financial service and insurance industry build machine learning solutions on AWS. In his spare time, he likes reading and teaching.