This post is co-authored by Anatoly Khomenko, Machine Learning Engineer, and Abdenour Bezzouh, Chief Technology Officer at Talent.com.
Established in 2011, Talent.com aggregates paid job listings from their clients and public job listings, and has created a unified, easily searchable platform. Covering over 30 million job listings across more than 75 countries and spanning various languages, industries, and distribution channels, Talent.com caters to the diverse needs of job seekers, effectively connecting millions of job seekers with job opportunities.
Talent.com’s mission is to facilitate global workforce connections. To achieve this, Talent.com aggregates job listings from various sources on the web, offering job seekers access to an extensive pool of over 30 million job opportunities tailored to their skills and experiences. In line with this mission, Talent.com collaborated with AWS to develop a cutting-edge job recommendation engine driven by deep learning, aimed at assisting users in advancing their careers.
To ensure the effective operation of this job recommendation engine, it is crucial to implement a large-scale data processing pipeline responsible for extracting and refining features from Talent.com’s aggregated job listings. This pipeline is able to process 5 million daily records in less than 1 hour, and allows for processing multiple days of records in parallel. In addition, this solution allows for a quick deployment to production. The primary source of data for this pipeline is the JSON Lines format, stored in Amazon Simple Storage Service (Amazon S3) and partitioned by date. Each day, this results in the generation of tens of thousands of JSON Lines files, with incremental updates occurring daily.
The primary objective of this data processing pipeline is to facilitate the creation of features necessary for training and deploying the job recommendation engine on Talent.com. It’s worth noting that this pipeline must support incremental updates and cater to the intricate feature extraction requirements necessary for the training and deployment modules essential for the job recommendation system. Our pipeline belongs to the general ETL (extract, transform, and load) process family that combines data from multiple sources into a large, central repository.
For further insights into how Talent.com and AWS collaboratively built cutting-edge natural language processing and deep learning model training techniques, utilizing Amazon SageMaker to craft a job recommendation system, refer to From text to dream job: Building an NLP-based job recommender at Talent.com with Amazon SageMaker. The system includes feature engineering, deep learning model architecture design, hyperparameter optimization, and model evaluation, where all modules are run using Python.
This post shows how we used SageMaker to build a large-scale data processing pipeline for preparing features for the job recommendation engine at Talent.com. The resulting solution enables a Data Scientist to ideate feature extraction in a SageMaker notebook using Python libraries, such as Scikit-Learn or PyTorch, and then to quickly deploy the same code into the data processing pipeline performing feature extraction at scale. The solution does not require porting the feature extraction code to use PySpark, as required when using AWS Glue as the ETL solution. Our solution can be developed and deployed solely by a Data Scientist end-to-end using only a SageMaker, and does not require knowledge of other ETL solutions, such as AWS Batch. This can significantly shorten the time needed to deploy the Machine Learning (ML) pipeline to production. The pipeline is operated through Python and seamlessly integrates with feature extraction workflows, rendering it adaptable to a wide range of data analytics applications.
Solution overview

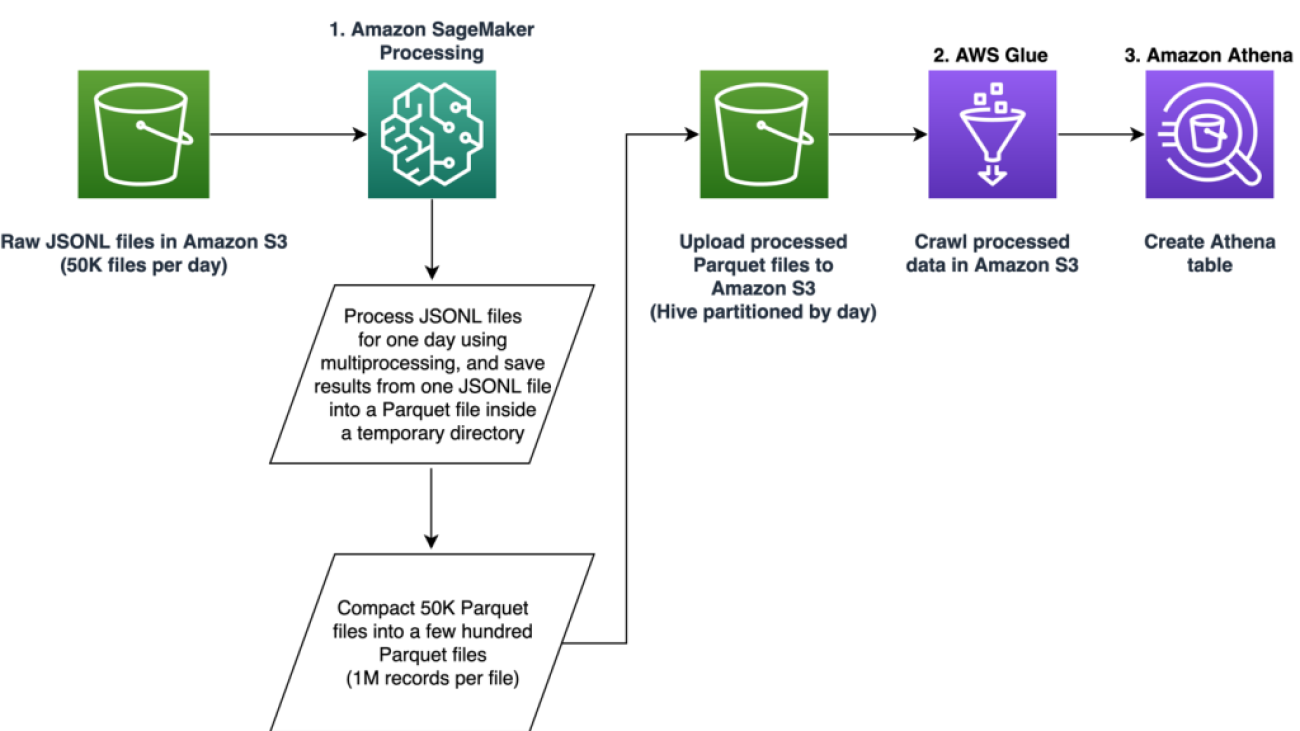
The pipeline is comprised of three primary phases:
- Utilize an Amazon SageMaker Processing job to handle raw JSONL files associated with a specified day. Multiple days of data can be processed by separate Processing jobs simultaneously.
- Employ AWS Glue for data crawling after processing multiple days of data.
- Load processed features for a specified date range using SQL from an Amazon Athena table, then train and deploy the job recommender model.
Process raw JSONL files
We process raw JSONL files for a specified day using a SageMaker Processing job. The job implements feature extraction and data compaction, and saves processed features into Parquet files with 1 million records per file. We take advantage of CPU parallelization to perform feature extraction for each raw JSONL file in parallel. Processing results of each JSONL file is saved into a separate Parquet file inside a temporary directory. After all of the JSONL files have been processed, we perform compaction of thousands of small Parquet files into several files with 1 million records per file. The compacted Parquet files are then uploaded into Amazon S3 as the output of the processing job. The data compaction ensures efficient crawling and SQL queries in the next stages of the pipeline.
The following is the sample code to schedule a SageMaker Processing job for a specified day, for example 2020-01-01, using the SageMaker SDK. The job reads raw JSONL files from Amazon S3 (for example from s3://bucket/raw-data/2020/01/01) and saves the compacted Parquet files into Amazon S3 (for example to s3://bucket/processed/table-name/day_partition=2020-01-01/).
### install dependencies
%pip install sagemaker pyarrow s3fs awswrangler
import sagemaker
import boto3
from sagemaker.processing import FrameworkProcessor
from sagemaker.sklearn.estimator import SKLearn
from sagemaker import get_execution_role
from sagemaker.processing import ProcessingInput, ProcessingOutput
region = boto3.session.Session().region_name
role = get_execution_role()
bucket = sagemaker.Session().default_bucket()
### we use instance with 16 CPUs and 128 GiB memory
### note that the script will NOT load the entire data into memory during compaction
### depending on the size of individual jsonl files, larger instance may be needed
instance = "ml.r5.4xlarge"
n_jobs = 8 ### we use 8 process workers
date = "2020-01-01" ### process data for one day
est_cls = SKLearn
framework_version_str = "0.20.0"
### schedule processing job
script_processor = FrameworkProcessor(
role=role,
instance_count=1,
instance_type=instance,
estimator_cls=est_cls,
framework_version=framework_version_str,
volume_size_in_gb=500,
)
script_processor.run(
code="processing_script.py", ### name of the main processing script
source_dir="../src/etl/", ### location of source code directory
### our processing script loads raw jsonl files directly from S3
### this avoids long start-up times of the processing jobs,
### since raw data does not need to be copied into instance
inputs=[], ### processing job input is empty
outputs=[
ProcessingOutput(destination="s3://bucket/processed/table-name/",
source="/opt/ml/processing/output"),
],
arguments=[
### directory with job's output
"--output", "/opt/ml/processing/output",
### temporary directory inside instance
"--tmp_output", "/opt/ml/tmp_output",
"--n_jobs", str(n_jobs), ### number of process workers
"--date", date, ### date to process
### location with raw jsonl files in S3
"--path", "s3://bucket/raw-data/",
],
wait=False
)
The following code outline for the main script (processing_script.py) that runs the SageMaker Processing job is as follows:
import concurrent
import pyarrow.dataset as ds
import os
import s3fs
from pathlib import Path
### function to process raw jsonl file and save extracted features into parquet file
from process_data import process_jsonl
### parse command line arguments
args = parse_args()
### we use s3fs to crawl S3 input path for raw jsonl files
fs = s3fs.S3FileSystem()
### we assume raw jsonl files are stored in S3 directories partitioned by date
### for example: s3://bucket/raw-data/2020/01/01/
jsons = fs.find(os.path.join(args.path, *args.date.split('-')))
### temporary directory location inside the Processing job instance
tmp_out = os.path.join(args.tmp_output, f"day_partition={args.date}")
### directory location with job's output
out_dir = os.path.join(args.output, f"day_partition={args.date}")
### process individual jsonl files in parallel using n_jobs process workers
futures=[]
with concurrent.futures.ProcessPoolExecutor(max_workers=args.n_jobs) as executor:
for file in jsons:
inp_file = Path(file)
out_file = os.path.join(tmp_out, inp_file.stem + ".snappy.parquet")
### process_jsonl function reads raw jsonl file from S3 location (inp_file)
### and saves result into parquet file (out_file) inside temporary directory
futures.append(executor.submit(process_jsonl, file, out_file))
### wait until all jsonl files are processed
for future in concurrent.futures.as_completed(futures):
result = future.result()
### compact parquet files
dataset = ds.dataset(tmp_out)
if len(dataset.schema) > 0:
### save compacted parquet files with 1MM records per file
ds.write_dataset(dataset, out_dir, format="parquet",
max_rows_per_file=1024 * 1024)
Scalability is a key feature of our pipeline. First, multiple SageMaker Processing jobs can be used to process data for several days simultaneously. Second, we avoid loading the entire processed or raw data into memory at once, while processing each specified day of data. This enables the processing of data using instance types that can’t accommodate a full day’s worth of data in primary memory. The only requirement is that the instance type should be capable of loading N raw JSONL or processed Parquet files into memory simultaneously, with N being the number of process workers in use.
Crawl processed data using AWS Glue
After all the raw data for multiple days has been processed, we can create an Athena table from the entire dataset by using an AWS Glue crawler. We use the AWS SDK for pandas (awswrangler) library to create the table using the following snippet:
import awswrangler as wr
### crawl processed data in S3
res = wr.s3.store_parquet_metadata(
path='s3://bucket/processed/table-name/',
database="database_name",
table="table_name",
dataset=True,
mode="overwrite",
sampling=1.0,
path_suffix='.parquet',
)
### print table schema
print(res[0])
Load processed features for training
Processed features for a specified date range can now be loaded from the Athena table using SQL, and these features can then be used for training the job recommender model. For example, the following snippet loads one month of processed features into a DataFrame using the awswrangler library:
import awswrangler as wr
query = """
SELECT *
FROM table_name
WHERE day_partition BETWEN '2020-01-01' AND '2020-02-01'
"""
### load 1 month of data from database_name.table_name into a DataFrame
df = wr.athena.read_sql_query(query, database='database_name')
Additionally, the use of SQL for loading processed features for training can be extended to accommodate various other use cases. For instance, we can apply a similar pipeline to maintain two separate Athena tables: one for storing user impressions and another for storing user clicks on these impressions. Using SQL join statements, we can retrieve impressions that users either clicked on or didn’t click on and then pass these impressions to a model training job.
Solution benefits
Implementing the proposed solution brings several advantages to our existing workflow, including:
- Simplified implementation – The solution enables feature extraction to be implemented in Python using popular ML libraries. And, it does not require the code to be ported into PySpark. This streamlines feature extraction as the same code developed by a Data Scientist in a notebook will be executed by this pipeline.
- Quick path-to-production – The solution can be developed and deployed by a Data Scientist to perform feature extraction at scale, enabling them to develop an ML recommender model against this data. At the same time, the same solution can be deployed to production by an ML Engineer with little modifications needed.
- Reusability – The solution provides a reusable pattern for feature extraction at scale, and can be easily adapted for other use cases beyond building recommender models.
- Efficiency – The solution offers good performance: processing a single day of the Talent.com’s data took less than 1 hour.
- Incremental updates – The solution also supports incremental updates. New daily data can be processed with a SageMaker Processing job, and the S3 location containing the processed data can be recrawled to update the Athena table. We can also use a cron job to update today’s data several times per day (for example, every 3 hours).
We used this ETL pipeline to help Talent.com process 50,000 files per day containing 5 million records, and created training data using features extracted from 90 days of raw data from Talent.com—a total of 450 million records across 900,000 files. Our pipeline helped Talent.com build and deploy the recommendation system into production within only 2 weeks. The solution performed all ML processes including ETL on Amazon SageMaker without utilizing other AWS service. The job recommendation system drove an 8.6% increase in clickthrough rate in online A/B testing against a previous XGBoost-based solution, helping connect millions of Talent.com’s users to better jobs.
Conclusion
This post outlines the ETL pipeline we developed for feature processing for training and deploying a job recommender model at Talent.com. Our pipeline uses SageMaker Processing jobs for efficient data processing and feature extraction at a large scale. Feature extraction code is implemented in Python enabling the use of popular ML libraries to perform feature extraction at scale, without the need to port the code to use PySpark.
We encourage the readers to explore the possibility of using the pipeline presented in this blog as a template for their use-cases where feature extraction at scale is required. The pipeline can be leveraged by a Data Scientist to build an ML model, and the same pipeline can then be adopted by an ML Engineer to run in production. This can significantly reduce the time needed to productize the ML solution end-to-end, as was the case with Talent.com. The readers can refer to the tutorial for setting up and running SageMaker Processing jobs. We also refer the readers to view the post From text to dream job: Building an NLP-based job recommender at Talent.com with Amazon SageMaker, where we discuss deep learning model training techniques utilizing Amazon SageMaker to build Talent.com’s job recommendation system.
About the authors
 Dmitriy Bespalov is a Senior Applied Scientist at the Amazon Machine Learning Solutions Lab, where he helps AWS customers across different industries accelerate their AI and cloud adoption.
Dmitriy Bespalov is a Senior Applied Scientist at the Amazon Machine Learning Solutions Lab, where he helps AWS customers across different industries accelerate their AI and cloud adoption.
 Yi Xiang is a Applied Scientist II at the Amazon Machine Learning Solutions Lab, where she helps AWS customers across different industries accelerate their AI and cloud adoption.
Yi Xiang is a Applied Scientist II at the Amazon Machine Learning Solutions Lab, where she helps AWS customers across different industries accelerate their AI and cloud adoption.
 Tong Wang is a Senior Applied Scientist at the Amazon Machine Learning Solutions Lab, where he helps AWS customers across different industries accelerate their AI and cloud adoption.
Tong Wang is a Senior Applied Scientist at the Amazon Machine Learning Solutions Lab, where he helps AWS customers across different industries accelerate their AI and cloud adoption.
 Anatoly Khomenko is a Senior Machine Learning Engineer at Talent.com with a passion for natural language processing matching good people to good jobs.
Anatoly Khomenko is a Senior Machine Learning Engineer at Talent.com with a passion for natural language processing matching good people to good jobs.
 Abdenour Bezzouh is an executive with more than 25 years experience building and delivering technology solutions that scale to millions of customers. Abdenour held the position of Chief Technology Officer (CTO) at Talent.com when the AWS team designed and executed this particular solution for Talent.com.
Abdenour Bezzouh is an executive with more than 25 years experience building and delivering technology solutions that scale to millions of customers. Abdenour held the position of Chief Technology Officer (CTO) at Talent.com when the AWS team designed and executed this particular solution for Talent.com.
 Yanjun Qi is a Senior Applied Science Manager at the Amazon Machine Learning Solution Lab. She innovates and applies machine learning to help AWS customers speed up their AI and cloud adoption.
Yanjun Qi is a Senior Applied Science Manager at the Amazon Machine Learning Solution Lab. She innovates and applies machine learning to help AWS customers speed up their AI and cloud adoption.














 NVIDIA GeForce NOW (@NVIDIAGFN)
NVIDIA GeForce NOW (@NVIDIAGFN)