Posted by Dave Steiner, Clinical Research Scientist, Google Health, and Rory Pilgrim, Product Manager Google Research

There’s a worldwide shortage of access to medical imaging expert interpretation across specialties including radiology, dermatology and pathology. Machine learning (ML) technology can help ease this burden by powering tools that enable doctors to interpret these images more accurately and efficiently. However, the development and implementation of such ML tools are often limited by the availability of high-quality data, ML expertise, and computational resources.
One way to catalyze the use of ML for medical imaging is via domain-specific models that utilize deep learning (DL) to capture the information in medical images as compressed numerical vectors (called embeddings). These embeddings represent a type of pre-learned understanding of the important features in an image. Identifying patterns in the embeddings reduces the amount of data, expertise, and compute needed to train performant models as compared to working with high-dimensional data, such as images, directly. Indeed, these embeddings can be used to perform a variety of downstream tasks within the specialized domain (see animated graphic below). This framework of leveraging pre-learned understanding to solve related tasks is similar to that of a seasoned guitar player quickly learning a new song by ear. Because the guitar player has already built up a foundation of skill and understanding, they can quickly pick up the patterns and groove of a new song.
 |
| Path Foundation is used to convert a small dataset of (image, label) pairs into (embedding, label) pairs. These pairs can then be used to train a task-specific classifier using a linear probe, (i.e., a lightweight linear classifier) as represented in this graphic, or other types of models using the embeddings as input. |
 |
| Once the linear probe is trained, it can be used to make predictions on embeddings from new images. These predictions can be compared to ground truth information in order to evaluate the linear probe’s performance. |
In order to make this type of embedding model available and drive further development of ML tools in medical imaging, we are excited to release two domain-specific tools for research use: Derm Foundation and Path Foundation. This follows on the strong response we’ve already received from researchers using the CXR Foundation embedding tool for chest radiographs and represents a portion of our expanding research offerings across multiple medical-specialized modalities. These embedding tools take an image as input and produce a numerical vector (the embedding) that is specialized to the domains of dermatology and digital pathology images, respectively. By running a dataset of chest X-ray, dermatology, or pathology images through the respective embedding tool, researchers can obtain embeddings for their own images, and use these embeddings to quickly develop new models for their applications.
Path Foundation
In “Domain-specific optimization and diverse evaluation of self-supervised models for histopathology”, we showed that self-supervised learning (SSL) models for pathology images outperform traditional pre-training approaches and enable efficient training of classifiers for downstream tasks. This effort focused on hematoxylin and eosin (H&E) stained slides, the principal tissue stain in diagnostic pathology that enables pathologists to visualize cellular features under a microscope. The performance of linear classifiers trained using the output of the SSL models matched that of prior DL models trained on orders of magnitude more labeled data.
Due to substantial differences between digital pathology images and “natural image” photos, this work involved several pathology-specific optimizations during model training. One key element is that whole-slide images (WSIs) in pathology can be 100,000 pixels across (thousands of times larger than typical smartphone photos) and are analyzed by experts at multiple magnifications (zoom levels). As such, the WSIs are typically broken down into smaller tiles or patches for computer vision and DL applications. The resulting images are information dense with cells or tissue structures distributed throughout the frame instead of having distinct semantic objects or foreground vs. background variations, thus creating unique challenges for robust SSL and feature extraction. Additionally, physical (e.g., cutting) and chemical (e.g., fixing and staining) processes used to prepare the samples can influence image appearance dramatically.
Taking these important aspects into consideration, pathology-specific SSL optimizations included helping the model learn stain-agnostic features, generalizing the model to patches from multiple magnifications, augmenting the data to mimic scanning and image post processing, and custom data balancing to improve input heterogeneity for SSL training. These approaches were extensively evaluated using a broad set of benchmark tasks involving 17 different tissue types over 12 different tasks.
Utilizing the vision transformer (ViT-S/16) architecture, Path Foundation was selected as the best performing model from the optimization and evaluation process described above (and illustrated in the figure below). This model thus provides an important balance between performance and model size to enable valuable and scalable use in generating embeddings over the many individual image patches of large pathology WSIs.
 |
| SSL training with pathology-specific optimizations for Path Foundation. |
The value of domain-specific image representations can also be seen in the figure below, which shows the linear probing performance improvement of Path Foundation (as measured by AUROC) compared to traditional pre-training on natural images (ImageNet-21k). This includes evaluation for tasks such as metastatic breast cancer detection in lymph nodes, prostate cancer grading, and breast cancer grading, among others.
 |
| Path Foundation embeddings significantly outperform traditional ImageNet embeddings as evaluated by linear probing across multiple evaluation tasks in histopathology. |
Derm Foundation
Derm Foundation is an embedding tool derived from our research in applying DL to interpret images of dermatology conditions and includes our recent work that adds improvements to generalize better to new datasets. Due to its dermatology-specific pre-training it has a latent understanding of features present in images of skin conditions and can be used to quickly develop models to classify skin conditions. The model underlying the API is a BiT ResNet-101×3 trained in two stages. The first pre-training stage uses contrastive learning, similar to ConVIRT, to train on a large number of image-text pairs from the internet. In the second stage, the image component of this pre-trained model is then fine-tuned for condition classification using clinical datasets, such as those from teledermatology services.
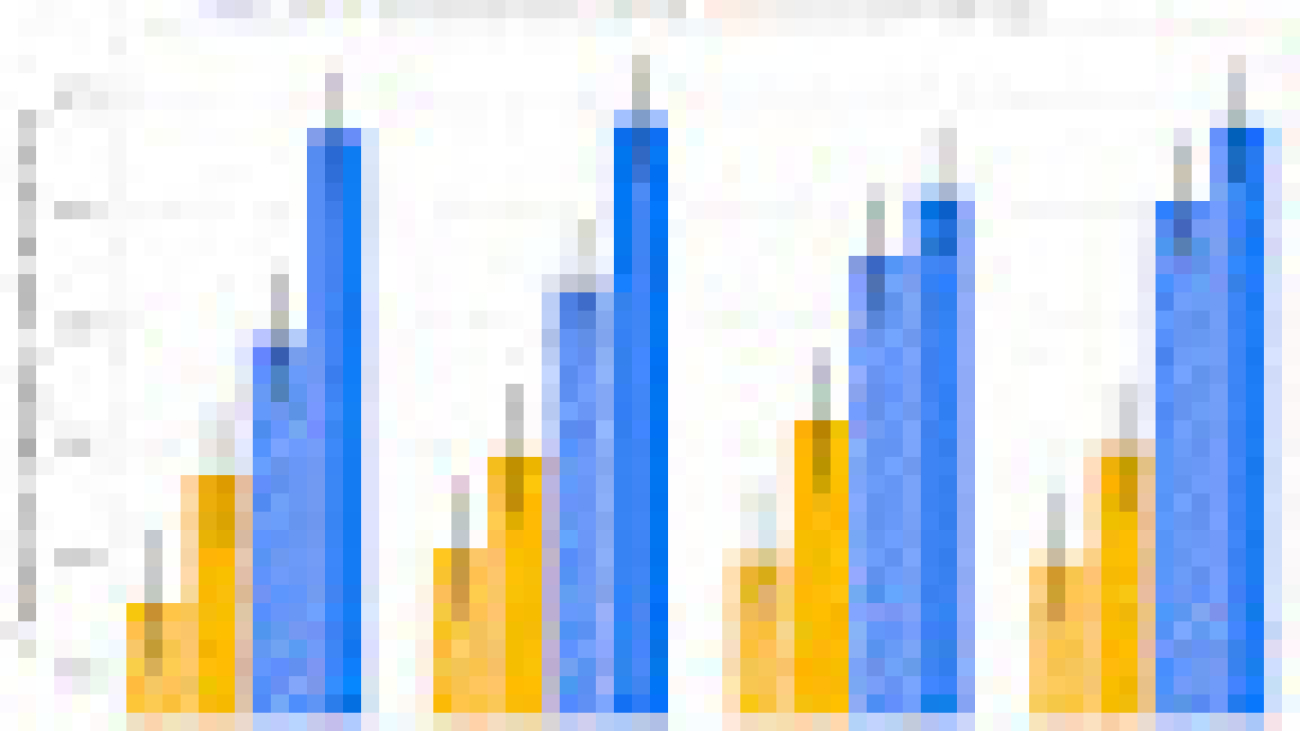
Unlike histopathology images, dermatology images more closely resemble the real-world images used to train many of today’s computer vision models. However, for specialized dermatology tasks, creating a high-quality model may still require a large dataset. With Derm Foundation, researchers can use their own smaller dataset to retrieve domain-specific embeddings, and use those to build smaller models (e.g., linear classifiers or other small non-linear models) that enable them to validate their research or product ideas. To evaluate this approach, we trained models on a downstream task using teledermatology data. Model training involved varying dataset sizes (12.5%, 25%, 50%, 100%) to compare embedding-based linear classifiers against fine-tuning.
The modeling variants considered were:
- A linear classifier on frozen embeddings from BiT-M (a standard pre-trained image model)
- Fine-tuned version of BiT-M with an extra dense layer for the downstream task
- A linear classifier on frozen embeddings from the Derm Foundation API
- Fine-tuned version of the model underlying the Derm Foundation API with an extra layer for the downstream task
We found that models built on top of the Derm Foundation embeddings for dermatology-related tasks achieved significantly higher quality than those built solely on embeddings or fine tuned from BiT-M. This advantage was found to be most pronounced for smaller training dataset sizes.
 |
| These results demonstrate that the Derm Foundation tooI can serve as a useful starting point to accelerate skin-related modeling tasks. We aim to enable other researchers to build on the underlying features and representations of dermatology that the model has learned. |
However, there are limitations with this analysis. We’re still exploring how well these embeddings generalize across task types, patient populations, and image settings. Downstream models built using Derm Foundation still require careful evaluation to understand their expected performance in the intended setting.
Access Path and Derm Foundation
We envision that the Derm Foundation and Path Foundation embedding tools will enable a range of use cases, including efficient development of models for diagnostic tasks, quality assurance and pre-analytical workflow improvements, image indexing and curation, and biomarker discovery and validation. We are releasing both tools to the research community so they can explore the utility of the embeddings for their own dermatology and pathology data.
To get access, please sign up to each tool’s terms of service using the following Google Forms.
After gaining access to each tool, you can use the API to retrieve embeddings from dermatology images or digital pathology images stored in Google Cloud. Approved users who are just curious to see the model and embeddings in action can use the provided example Colab notebooks to train models using public data for classifying six common skin conditions or identifying tumors in histopathology patches. We look forward to seeing the range of use-cases these tools can unlock.
Acknowledgements
We would like to thank the many collaborators who helped make this work possible including Yun Liu, Can Kirmizi, Fereshteh Mahvar, Bram Sterling, Arman Tajback, Kenneth Philbrik, Arnav Agharwal, Aurora Cheung, Andrew Sellergren, Boris Babenko, Basil Mustafa, Jan Freyberg, Terry Spitz, Yuan Liu, Pinal Bavishi, Ayush Jain, Amit Talreja, Rajeev Rikhye, Abbi Ward, Jeremy Lai, Faruk Ahmed, Supriya Vijay,Tiam Jaroensri, Jessica Loo, Saurabh Vyawahare, Saloni Agarwal, Ellery Wulczyn, Jonathan Krause, Fayaz Jamil, Tom Small, Annisah Um’rani, Lauren Winer, Sami Lachgar, Yossi Matias, Greg Corrado, and Dale Webster.
Read More










 For International Women’s day, we’re highlighting AI solutions that can improve early diagnosis, quality and access to care for women’s health worldwide.
For International Women’s day, we’re highlighting AI solutions that can improve early diagnosis, quality and access to care for women’s health worldwide.