As machine learning (ML) applications become more popular, customers are looking to streamline the process for developing, deploying, and continuously improving models. To reliably increase the frequency and quality of this cycle, customers are turning to ML operations (MLOps), which is the discipline of bringing continuous delivery principles and practices to the data science team. The following diagram illustrates the continuous deployment workflow.
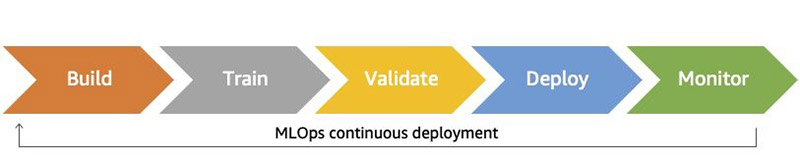
There are many ways to operationalizing ML. In this post, you see how to build an ML model that predicts taxi fares in New York City using Amazon SageMaker, AWS CodePipeline, and AWS CodeDeploy in a safe blue/green deployment pipeline.
Amazon SageMaker is a fully managed service that provides developers and data scientists the ability to quickly build, train, deploy, and monitor ML models. When combined with CodePipeline and CodeDeploy, it’s easy to create a fully serverless build pipeline with best practices in security and performance with lower costs.
CodePipeline is a fully managed continuous delivery service that helps you automate your release pipelines for fast and reliable application and infrastructure updates. CodePipeline automates the Build, Test, and Deploy phases of your release process every time a code change occurs, based on the release model you define.
What is blue/green deployment?
Blue/green deployment is a technique that reduces downtime and risk by running two identical production environments called Blue and Green. After you deploy a fully tested model to the Green endpoint, a fraction of traffic (for this use case, 10%) is sent to this new replacement endpoint. This continues for a period of time while there are no errors with an optional ramp up to reach 100% of traffic, at which point the Blue endpoint can be decommissioned. Green becomes live, and the process can repeat again. If any errors are detected during this process, a rollback occurs; Blue remains live and Green is decommissioned. The following diagram illustrates this architecture.
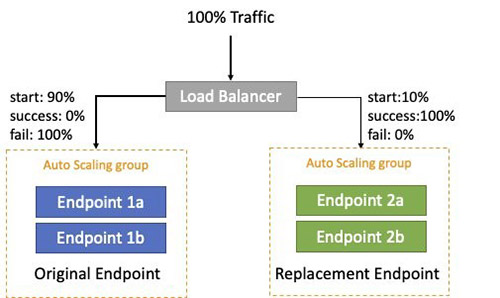
In this solution, the blue/green deployment is managed by AWS CodeDeploy with the AWS Lambda compute platform to switch between the blue/green autoscaling Amazon SageMaker endpoints.
Solution overview
For this post, you use a publicly available New York green taxi dataset to train an ML model to predict the fare amount using the Amazon SageMaker built-in XGBoost algorithm.
You automate the process of training, deploying, and monitoring the model with CodePipeline, which you orchestrate within an Amazon SageMaker notebook.
Getting started
This walkthrough uses AWS CloudFormation to create a continuous integration pipeline. You can configure this to a public or private GitHub repo with your own access token, or you can use an AWS CodeCommit repository in your environment that is cloned from the public GitHub repo.
Complete the following steps:
- Optionally, fork a copy of the GitHub repo into your own GitHub account by choosing the fork
- Create a personal access token (OAuth 2) with the scopes (permissions)
admin:repo_hook and repo. If you already have a token with these permissions, you can use that. You can find a list of all your personal access tokens in https://github.com/settings/tokens.
- Copy the access token to your clipboard. For security reasons, after you navigate off the page, you can’t see the token again. If you lose your token, you can regenerate
- Choose Launch Stack:

- Enter the following parameters:
- Model Name – A unique name for this model (must be fewer than 15 characters)
- Notebook Instance Type – The Amazon SageMaker instance type (default is ml.t3.medium)
- GitHub Access Token – Your access token
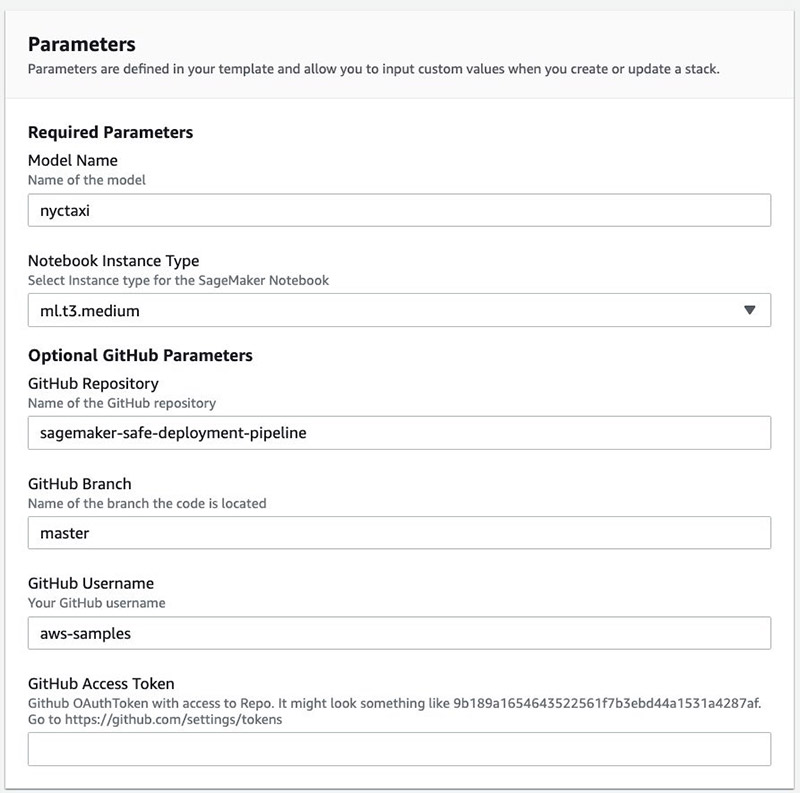
- Acknowledge that AWS CloudFormation may create additional AWS Identity and Access Management (IAM) resources.
- Choose Create stack.
The CloudFormation template creates an Amazon SageMaker notebook and pipeline.
When the deployment is complete, you have a new pipeline linked to your GitHub source. It starts in a Failed state because it’s waiting on an Amazon Simple Storage Service (Amazon S3) data source.
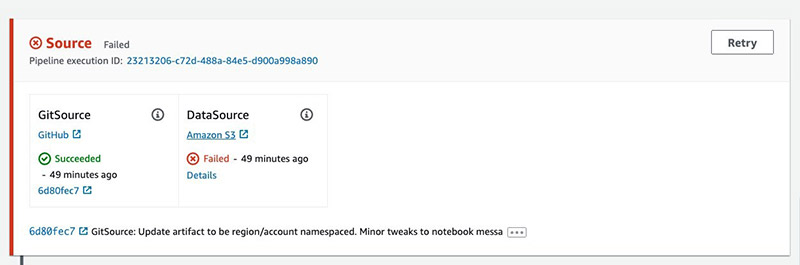
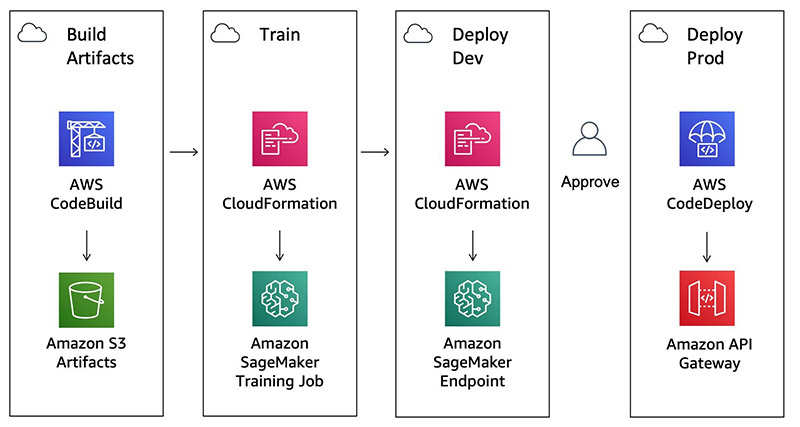
The pipeline has the following stages:
- Build Artifacts – Run a CodeBuild job to create CloudFormation templates.
- Train – Train an Amazon SageMaker pipeline and baseline processing job.
- Deploy Dev – Deploys a development Amazon SageMaker endpoint.
- Manual Approval – The user gives approval.
- Deploy Prod – Deploys an Amazon API Gateway AWS Lambda function in front of the Amazon SageMaker endpoints using CodeDeploy for blue/green deployment and rollback.
The following diagram illustrates this workflow.
Running the pipeline
Launch the newly created Amazon SageMaker notebook in your AWS account. For more information, see Build, Train, and Deploy a Machine Learning Model.
Navigate to the notebook directory and open the notebook by choosing the mlops.ipynb link.

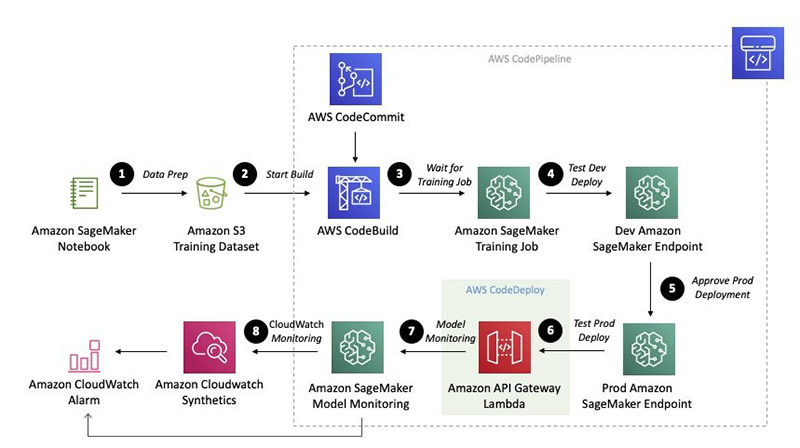
The notebook guides you through a series of steps, which we also review in this post:
- Data Prep
- Start Build
- Wait for Training Job
- Test Dev Deployment
- Approve Prod Deployment
- Test Prod Deployment
- Model Monitoring
- CloudWatch Monitoring
The following diagram illustrates this workflow.
Step 1: Data Prep
In this step, you download the February 2018 trips from New York green taxi trip records to a local file for input into a pandas DataFrame. See the following code:
import pandas as pd
parse_dates = ["lpep_dropoff_datetime", "lpep_pickup_datetime"]
trip_df = pd.read_csv("nyc-tlc.csv", parse_dates=parse_dates)
You then add a feature engineering step to calculate the duration in minutes from the pick-up and drop-off times:
trip_df["duration_minutes"] = (
trip_df["lpep_dropoff_datetime"] - trip_df["lpep_pickup_datetime"]
).dt.seconds / 60
You create a new DataFrame just to include the total amount as the target column, using duration in minutes, passenger count, and trip distance as input features:
cols = ["total_amount", "duration_minutes", "passenger_count", "trip_distance"]
data_df = trip_df[cols]
print(data_df.shape)
data_df.head()
The following table shows you the first five rows in your DataFrame.
|
total_amount |
duration_minutes |
passenger_count |
trip_distance |
| 1 |
23 |
0.05 |
1 |
0 |
| 2 |
9.68 |
7.11667 |
5 |
1.6 |
| 3 |
35.76 |
22.81667 |
1 |
9.6 |
| 4 |
5.8 |
3.16667 |
1 |
0.73 |
| 5 |
9.3 |
6.63333 |
2 |
1.87 |
Continue through the notebook to visualize a sample of the DataFrame, before splitting the dataset into 80% training, 15% validation, and 5% test. See the following code:
from sklearn.model_selection import train_test_split
train_df, val_df = train_test_split(data_df, test_size=0.20, random_state=42)
val_df, test_df = train_test_split(val_df, test_size=0.05, random_state=42)
# Set the index for our test dataframe
test_df.reset_index(inplace=True, drop=True)
print('split train: {}, val: {}, test: {} '.format(train_df.shape[0], val_df.shape[0], test_df.shape[0]))
Step 2: Start Build
The pipeline source has two inputs:
- A Git source repository containing the model definition and all supporting infrastructure
- An Amazon S3 data source that includes a reference to the training and validation datasets
The Start Build section in the notebook uploads a .zip file to the Amazon S3 data source that triggers the build. See the following code:
from io import BytesIO
import zipfile
import json
input_data = {
"TrainingUri": s3_train_uri,
"ValidationUri": s3_val_uri,
"BaselineUri": s3_baseline_uri,
}
hyperparameters = {"num_round": 50}
data_source_key = "{}/data-source.zip".format(pipeline_name)
zip_buffer = BytesIO()
with zipfile.ZipFile(zip_buffer, "a") as zf:
zf.writestr("inputData.json", json.dumps(input_data))
zf.writestr("hyperparameters.json", json.dumps(hyperparameters))
zip_buffer.seek(0)
s3 = boto3.client("s3")
s3.put_object(
Bucket=artifact_bucket, Key=data_source_key, Body=bytearray(zip_buffer.read())
)
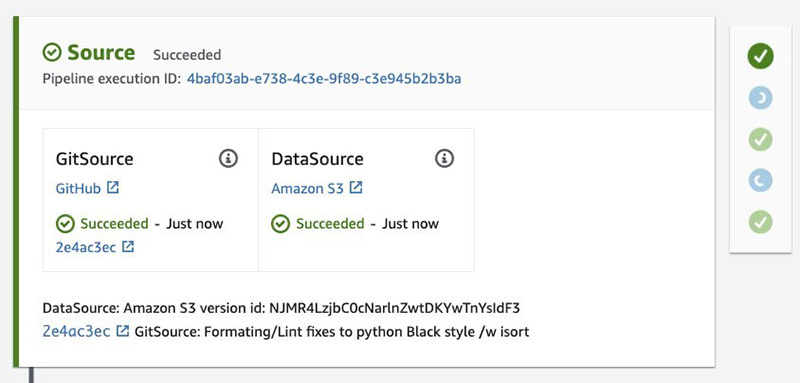
Specifically, you see a VersionId in the output from this cell:
{'ResponseMetadata': {'RequestId': 'ED389631CA6A9815',
'HostId': '3jAk/BJoRb78yElCVxrEpekVKE34j/WKIqwTIJIxgb2IoUSV8khz7T5GLiSKO/u0c66h8/Iye9w=',
'HTTPStatusCode': 200,
'HTTPHeaders': {'x-amz-id-2': '3jAk/BJoRb78yElCVxrEpekVKE34j/WKIqwTIJIxgb2IoUSV8khz7T5GLiSKO/u0c66h8/Iye9w=',
'x-amz-request-id': 'ED389631CA6A9815',
'date': 'Mon, 15 Jun 2020 05:06:39 GMT',
'x-amz-version-id': 'NJMR4LzjbC0cNarlnZwtDKYwTnYsIdF3',
'etag': '"47f9ca2b44d0e2d66def2f098dd13094"',
'content-length': '0',
'server': 'AmazonS3'},
'RetryAttempts': 0},
'ETag': '"47f9ca2b44d0e2d66def2f098dd13094"',
'VersionId': 'NJMR4LzjbC0cNarlnZwtDKYwTnYsIdF3'}
This corresponds to the Amazon S3 data source version id in the pipeline. See the following screenshot.
The Build stage in the pipeline runs a CodeBuild job defined in buildspec.yml that runs the following actions:
- Runs the model
run.py Python file to output the training job definition.
- Packages CloudFormation templates for the Dev and Prod deploy stages, including the API Gateway and Lambda resources for the blue/green deployment.
The source code for the model and API are available in the Git repository. See the following directory tree:
├── api
│ ├── __init__.py
│ ├── app.py
│ ├── post_traffic_hook.py
│ └── pre_traffic_hook.py
├── model
│ ├── buildspec.yml
│ ├── requirements.txt
│ └── run.py
The Build stage is also responsible for deploying the Lambda custom resources referenced in the CloudFormation stacks.
Step 3: Wait for Training Job
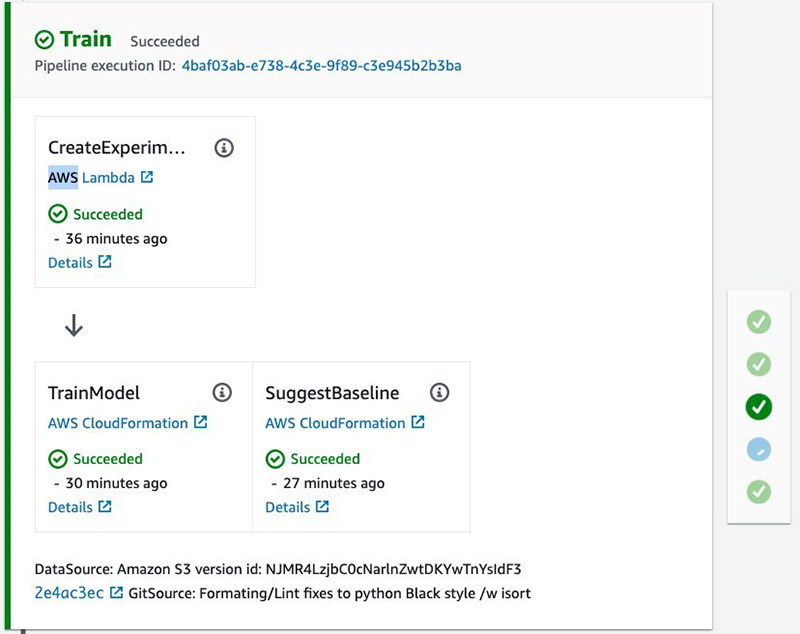
When the training and baseline job is complete, you can inspect the metrics associated with the Experiment and Trial component linked to the pipeline execution ID.
In addition to the train metrics, there is another job to create a baseline, which outputs statistics and constraints used for model monitoring after the model is deployed to production. The following table summarizes the parameters.
| TrialComponentName |
DisplayName |
SageMaker.InstanceType |
train:rmse – Last |
validation:rmse – Last |
| mlops-nyctaxi-pbl-4baf03ab-e738-4c3e-9f89-c3e945b2b3ba-aws-processing-job |
Baseline |
ml.m5.xlarge |
NaN |
NaN |
| mlops-nyctaxi-4baf03ab-e738-4c3e-9f89-c3e945b2b3ba-aws-training-job |
Training |
ml.m4.xlarge |
2.69262 |
2.73961 |
These actions run in parallel using custom AWS CloudFormation resources that poll the jobs every 2 minutes to check on their status. See the following screenshot.
Step 4: Test Dev Deployment
With the training job complete, the development endpoint is deployed in the next stage. The notebook polls the endpoint status until it becomes InService. See the following code:
sm = boto3.client('sagemaker')
while True:
try:
response = sm.describe_endpoint(EndpointName=dev_endpoint_name)
print("Endpoint status: {}".format(response['EndpointStatus']))
if response['EndpointStatus'] == 'InService':
break
except:
pass
time.sleep(10)
With an endpoint in service, you can use the notebook to predict the expected fare amount based on the inputs from the test dataset. See the following code:
pred_df = pd.DataFrame({'total_amount_predictions': predictions })
pred_df = test_df.join(pred_df) # Join on all
pred_df['error'] = abs(pred_df['total_amount']-pred_df['total_amount_predictions'])
ax = pred_df.tail(1000).plot.scatter(x='total_amount_predictions', y='total_amount',
c='error', title='actual amount vs pred')
You can join these predictions back to the target total amount, and visualize them in a scatter plot.
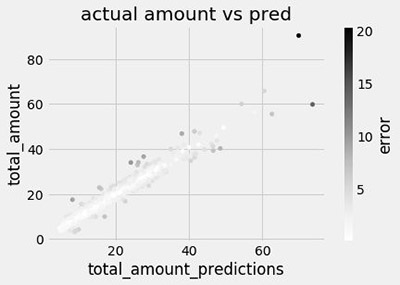
The notebook also calculates the root mean square error, which is commonly used in regression problems like this.
Step 5: Approve Prod Deployment
If you’re happy with the model, you can approve it directly using the Jupyter notebook widget.

As an administrator, you can also approve or reject the manual approval on the AWS CodePipeline console.
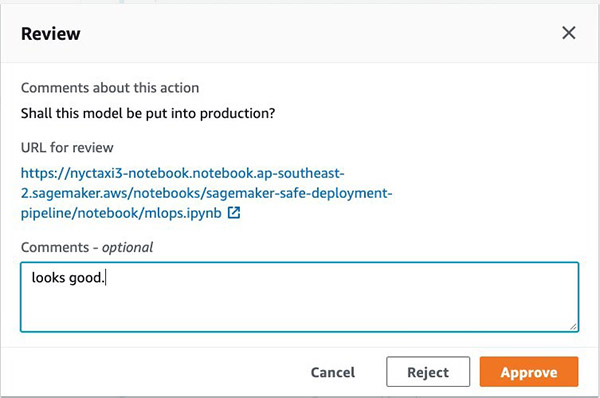
Approving this action moves the pipeline to the final blue/green production deployment stage.
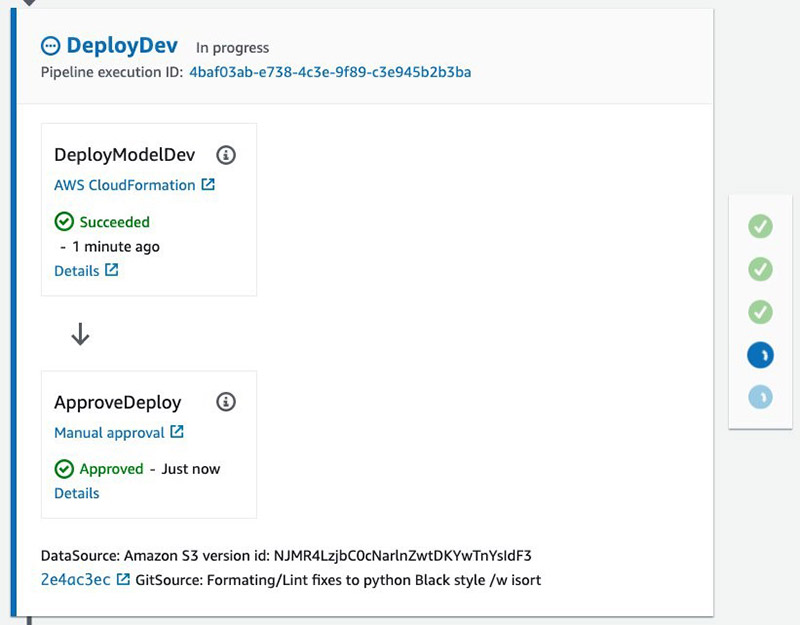
Step 6: Test Prod Deployment
Production deployment is managed through a single AWS CloudFormation which performs several dependent actions, including:
- Creates an Amazon SageMaker endpoint with AutoScaling enabled
- Updates the endpoints to enable data capture and schedule model monitoring
- Calls CodeDeploy to create or update a RESTful API using blue/green Lambda deployment
The following diagram illustrates this workflow.
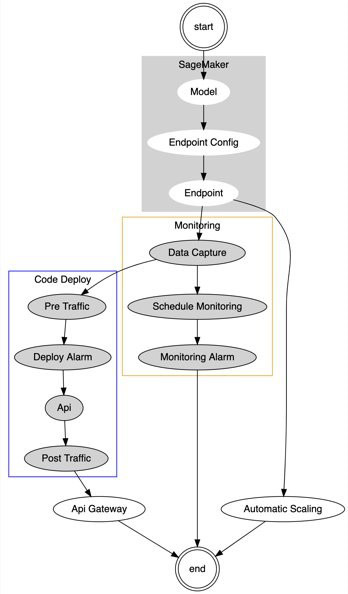
The first time this pipeline is run, there’s no existing Blue deployment, so CodeDeploy creates a new API Gateway and Lambda resource, which is configured to invoke an Amazon SageMaker endpoint that has been configured with AutoScaling and data capture enabled.
Rerunning the build pipeline
If you go back to Step 2 in the notebook and upload a new Amazon S3 data source artifact, you trigger a new build in the pipeline, which results in an update to the production deployment CloudFormation stack. This results in a new Lambda version pointing to a second Green Amazon SageMaker endpoint being created in parallel with the original Blue endpoint.
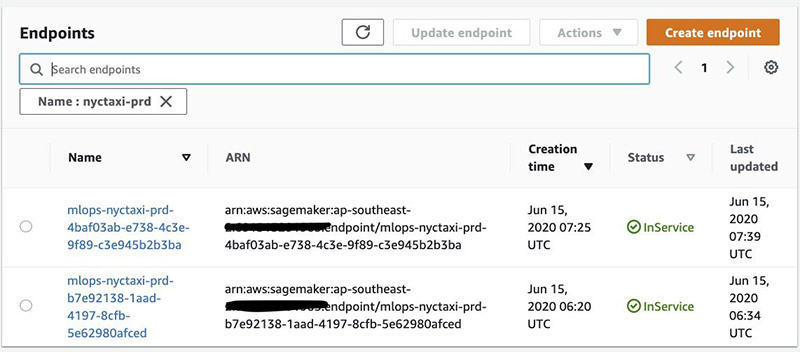
You can use the notebook to query the most recent events in the CloudFormation stack associated with this deployment. See the following code:
from datetime import datetime
from dateutil.tz import tzlocal
def get_event_dataframe(events):
stack_cols = [
"LogicalResourceId",
"ResourceStatus",
"ResourceStatusReason",
"Timestamp",
]
stack_event_df = pd.DataFrame(events)[stack_cols].fillna("")
stack_event_df["TimeAgo"] = datetime.now(tzlocal()) - stack_event_df["Timestamp"]
return stack_event_df.drop("Timestamp", axis=1)
# Get latest stack events
response = cfn.describe_stack_events(StackName=stack_name)
get_event_dataframe(response["StackEvents"]).head()
The output shows the latest events and, for this post, illustrates that the endpoint has been updated for data capture and CodeDeploy is in the process of switching traffic to the new endpoint. The following table summarizes the output.
| LogicalResourceId |
ResourceStatus |
ResourceStatusReason |
TimeAgo |
| PreTrafficLambdaFunction |
UPDATE_COMPLETE |
|
00:06:17.143352 |
| SagemakerDataCapture |
UPDATE_IN_PROGRESS |
|
00:06:17.898352 |
| PreTrafficLambdaFunction |
UPDATE_IN_PROGRESS |
|
00:06:18.114352 |
| Endpoint |
UPDATE_COMPLETE |
|
00:06:20.911352 |
| Endpoint |
UPDATE_IN_PROGRESS |
Resource creation Initiated |
00:12:56.016352 |
When the Blue endpoint status is InService, the notebook outputs a link to the CodeDeploy Deployment Application page, where you can watch as the traffic shifts from the original to the replacement endpoint.
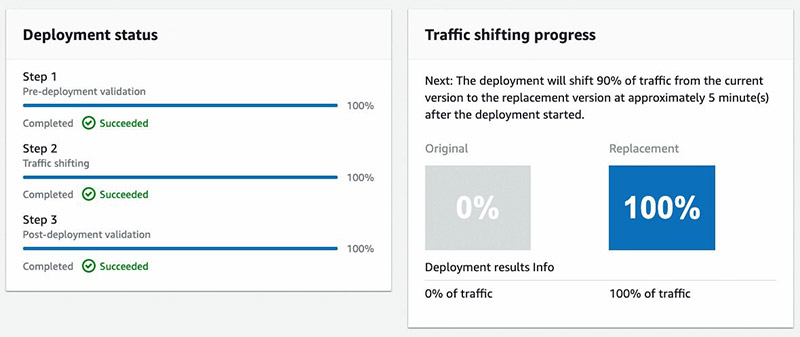
A successful blue/green deployment is contingent on the post-deployment validation passing, which for this post is configured to check that live traffic has been received; evident by data capture logs in Amazon S3.
The notebook guides you through the process of sending requests to the RESTful API, which is provided as an output in the CloudFormation stack. See the following code:
from urllib import request
headers = {"Content-type": "text/csv"}
payload = test_df[test_df.columns[1:]].head(1).to_csv(header=False, index=False).encode('utf-8')
while True:
try:
resp = request.urlopen(request.Request(outputs['RestApi'], data=payload, headers=headers))
print("Response code: %d: endpoint: %s" % (resp.getcode(), resp.getheader('x-sagemaker-endpoint')))
status, outputs = get_stack_status(stack_name)
if status.endswith('COMPLETE'):
print('Deployment completen')
break
except Exception as e:
pass
time.sleep(10)
This cell loops every 10 seconds until the deployment is complete, retrieving a header that indicates which Amazon SageMaker endpoint was hit for that request. Because we’re using the canary mode, you see a small sample of hits from the new target endpoint (ending in c3e945b2b3ba) until the CodeDeploy process completes successfully, at which point the original endpoint (ending in 5e62980afced) is deleted because it’s no longer required. See the following output:
Response code: 200: endpoint: mlops-nyctaxi-prd-b7e92138-1aad-4197-8cfb-5e62980afced
Response code: 200: endpoint: mlops-nyctaxi-prd-4baf03ab-e738-4c3e-9f89-c3e945b2b3ba
Response code: 200: endpoint: mlops-nyctaxi-prd-b7e92138-1aad-4197-8cfb-5e62980afced
Response code: 200: endpoint: mlops-nyctaxi-prd-b7e92138-1aad-4197-8cfb-5e62980afced
Response code: 200: endpoint: mlops-nyctaxi-prd-b7e92138-1aad-4197-8cfb-5e62980afced
Response code: 200: endpoint: mlops-nyctaxi-prd-b7e92138-1aad-4197-8cfb-5e62980afced
Response code: 200: endpoint: mlops-nyctaxi-prd-4baf03ab-e738-4c3e-9f89-c3e945b2b3ba
Response code: 200: endpoint: mlops-nyctaxi-prd-4baf03ab-e738-4c3e-9f89-c3e945b2b3ba
Response code: 200: endpoint: mlops-nyctaxi-prd-4baf03ab-e738-4c3e-9f89-c3e945b2b3ba
Response code: 200: endpoint: mlops-nyctaxi-prd-4baf03ab-e738-4c3e-9f89-c3e945b2b3ba
Response code: 200: endpoint: mlops-nyctaxi-prd-4baf03ab-e738-4c3e-9f89-c3e945b2b3ba
Response code: 200: endpoint: mlops-nyctaxi-prd-4baf03ab-e738-4c3e-9f89-c3e945b2b3ba
Response code: 200: endpoint: mlops-nyctaxi-prd-4baf03ab-e738-4c3e-9f89-c3e945b2b3ba
Step 7: Model Monitoring
As part of the production deployment, Amazon SageMaker Model Monitor is scheduled to run every hour on the newly created endpoint, which has been configured to capture data request input and output data to Amazon S3.
You can use the notebook to list these data capture files, which are collected as a series of JSON lines. See the following code:
bucket = sagemaker_session.default_bucket()
data_capture_logs_uri = 's3://{}/{}/datacapture/{}'.format(bucket, model_name, prd_endpoint_name)
capture_files = S3Downloader.list(data_capture_logs_uri)
print('Found {} files'.format(len(capture_files)))
if capture_files:
# Get the first line of the most recent file
event = json.loads(S3Downloader.read_file(capture_files[-1]).split('n')[0])
print('nLast file:n{}'.format(json.dumps(event, indent=2)))
If you take the first line of the last file, you can see that the input contains a CSV with fields for the following:
- Duration (10.65 minutes)
- Passenger count (1 person)
- Trip distance (2.56 miles)
The output is the following:
See the following code:
Found 8 files
Last file:
{
"captureData": {
"endpointInput": {
"observedContentType": "text/csv",
"mode": "INPUT",
"data": "10.65,1,2.56n",
"encoding": "CSV"
},
"endpointOutput": {
"observedContentType": "text/csv; charset=utf-8",
"mode": "OUTPUT",
"data": "12.720224380493164",
"encoding": "CSV"
}
},
"eventMetadata": {
"eventId": "44daf7d7-97c8-4504-8b3d-399891f8f217",
"inferenceTime": "2020-05-12T04:18:39Z"
},
"eventVersion": "0"
}
The monitoring job rolls up all the results in the last hour and compares these to the baseline statistics and constraints captured in the Train phase of the pipeline. As we can see from the notebook visualization, we can detect baseline drift with respect to the total_amount and trip_distance inputs, which were randomly sampled from our test set.

Step 8: CloudWatch Monitoring
AWS CloudWatch Synthetics provides you a way to set up a canary to test that your API is returning a successful HTTP 200 response on a regular interval. This is a great way to validate that the blue/green deployment isn’t causing any downtime for your end-users.
The notebook loads the canary.js template, which is parameterized with a rest_uri and payload invoked from the Lambda layer created by the canary, which is configured to hit the production REST API every 10 minutes. See the following code:
from urllib.parse import urlparse
from string import Template
from io import BytesIO
import zipfile
import sagemaker
# Format the canary_js with rest_api and payload
rest_url = urlparse(rest_api)
with open('canary.js') as f:
canary_js = Template(f.read()).substitute(hostname=rest_url.netloc,
path=rest_url.path,
data=payload.decode('utf-8').strip())
# Write the zip file
zip_buffer = BytesIO()
with zipfile.ZipFile(zip_buffer, 'w') as zf:
zip_path = 'nodejs/node_modules/apiCanaryBlueprint.js' # Set a valid path
zip_info = zipfile.ZipInfo(zip_path)
zip_info.external_attr = 0o0755 << 16 # Ensure the file is readable
zf.writestr(zip_info, canary_js)
zip_buffer.seek(0)
# Create the canary
synth = boto3.client('synthetics')
role = sagemaker.get_execution_role()
s3_canary_uri = 's3://{}/{}'.format(artifact_bucket, model_name)
canary_name = 'mlops-{}'.format(model_name)
response = synth.create_canary(
Name=canary_name,
Code={
'ZipFile': bytearray(zip_buffer.read()),
'Handler': 'apiCanaryBlueprint.handler'
},
ArtifactS3Location=s3_canary_uri,
ExecutionRoleArn=role,
Schedule={
'Expression': 'rate(10 minutes)',
'DurationInSeconds': 0 },
RunConfig={
'TimeoutInSeconds': 60,
'MemoryInMB': 960
},
SuccessRetentionPeriodInDays=31,
FailureRetentionPeriodInDays=31,
RuntimeVersion='syn-1.0',
)
You can also configure an alarm for this canary to alert when the success rates drop below 90%:
cloudwatch = boto3.client('cloudwatch')
canary_alarm_name = '{}-synth-lt-threshold'.format(canary_name)
response = cloudwatch.put_metric_alarm(
AlarmName=canary_alarm_name,
ComparisonOperator='LessThanThreshold',
EvaluationPeriods=1,
DatapointsToAlarm=1,
Period=600, # 10 minute interval
Statistic='Average',
Threshold=90.0,
ActionsEnabled=False,
AlarmDescription='SuccessPercent LessThanThreshold 90%',
Namespace='CloudWatchSynthetics',
MetricName='SuccessPercent',
Dimensions=[
{
'Name': 'CanaryName',
'Value': canary_name
},
],
Unit='Seconds'
)
With the canary created, you can choose the link provided to view the detail on the AWS CloudWatch console, which includes metrics such as uptime and as the logs output from your Lambda code.
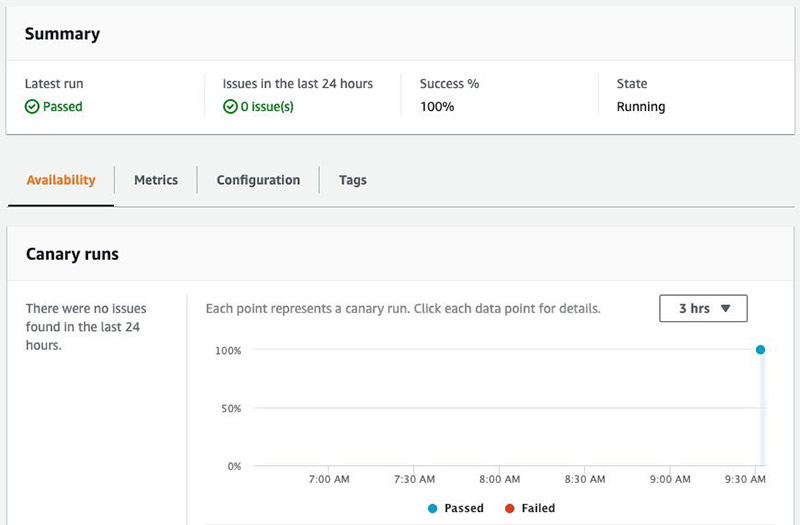
Returning to the notebook, create a CloudWatch dashboard from a template parametrized with the current region, account_id and model_name:
sts = boto3.client('sts')
account_id = sts.get_caller_identity().get('Account')
dashboard_name = 'mlops-{}'.format(model_name)
with open('dashboard.json') as f:
dashboard_body = Template(f.read()).substitute(region=region,
account_id=account_id,
model_name=model_name)
response = cloudwatch.put_dashboard(
DashboardName=dashboard_name,
DashboardBody=dashboard_body
)
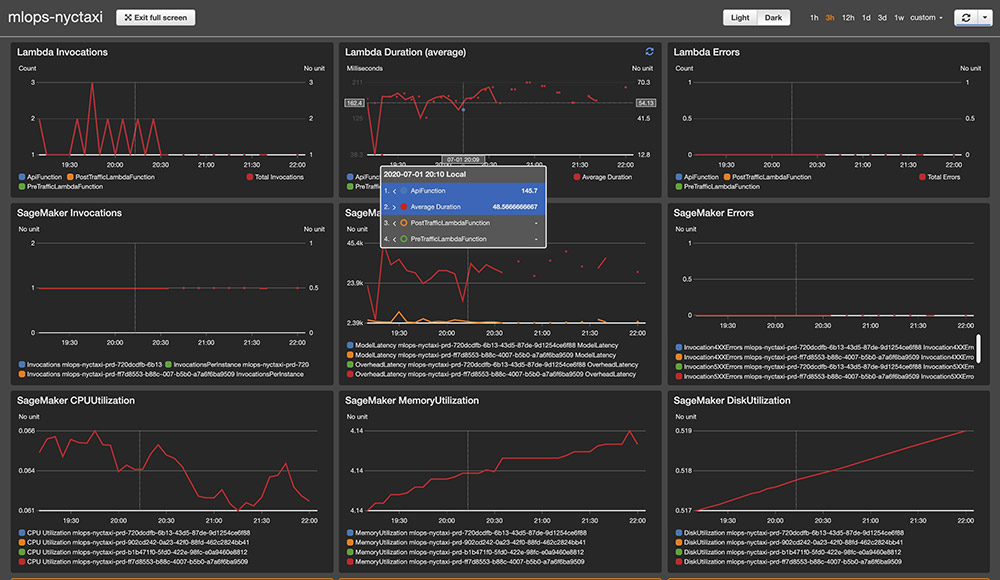
This creates a dashboard with a four-row-by-three-column layout with metrics for your production deployment, including the following:
- Lambda metrics for latency and throughput
- Amazon SageMaker endpoint
- CloudWatch alarms for CodeDeploy and model drift
You can choose the link to open the dashboard in full screen and use Dark mode. See the following screenshot.
Cleaning up
You can remove the canary and CloudWatch dashboard directly within the notebook. The pipeline created Amazon SageMaker training, baseline jobs, and endpoints using AWS CloudFormation, so to clean up these resources, delete the stacks prefixed with the name of your model. For example, for nyctaxi, the resources are the following:
- nyctaxi-devploy-prd
- nyctaxi-devploy-dev
- nyctaxi-training-job
- nyctaxi-suggest-baseline
After these are deleted, complete your cleanup by emptying the S3 bucket created to store your pipeline artefacts, and delete the original stack, which removes the pipeline, Amazon SageMaker notebook, and other resources.
Conclusion
In this post, we walked through creating an end-to-end safe deployment pipeline for Amazon SageMaker models using native AWS development tools CodePipeline, CodeBuild, and CodeDeploy. We demonstrated how you can trigger the pipeline directly from within an Amazon SageMaker notebook, validate and approve deployment to production, and continue to monitor your models after they go live. By running the pipeline a second time, we saw how CodeDeploy created a new Green replacement endpoint that was cut over to after the post-deployment validated it had received live traffic. Finally, we saw how to use CloudWatch Synthetics to constantly monitor our live endpoints and visualize metrics in a custom CloudWatch dashboard.
You could easily extend this solution to support your own dataset and model. The source code is available on the GitHub repo.
About the Author
 Julian Bright is an Sr. AI/ML Specialist Solutions Architect based out of Melbourne, Australia. Julian works as part of the global AWS machine learning team and is passionate about helping customers realise their AI and ML journey through MLOps. In his spare time he loves running around after his kids, playing soccer and getting outdoors.
Julian Bright is an Sr. AI/ML Specialist Solutions Architect based out of Melbourne, Australia. Julian works as part of the global AWS machine learning team and is passionate about helping customers realise their AI and ML journey through MLOps. In his spare time he loves running around after his kids, playing soccer and getting outdoors.
Read More


 Phi Nguyen is a solution architect at AWS helping customers with their cloud journey with a special focus on data lake, analytics, semantics technologies and machine learning. In his spare time, you can find him biking to work, coaching his son’s soccer team or enjoying nature walk with his family.
Phi Nguyen is a solution architect at AWS helping customers with their cloud journey with a special focus on data lake, analytics, semantics technologies and machine learning. In his spare time, you can find him biking to work, coaching his son’s soccer team or enjoying nature walk with his family. Xiang Song is an Applied Scientist with the AWS Shanghai AI Lab. He got his Bachelor’s degree in Software Engineer and Ph.D’s in Operating System and Architecture from Fudan University. His research interests include building machine learning systems and graph neural network for real world applications.
Xiang Song is an Applied Scientist with the AWS Shanghai AI Lab. He got his Bachelor’s degree in Software Engineer and Ph.D’s in Operating System and Architecture from Fudan University. His research interests include building machine learning systems and graph neural network for real world applications.



























 Amit Choudhary is a Senior Product Manager Technical Intern. He loves to build products that help developers learn about various machine learning techniques in a fun and hands-on manner.
Amit Choudhary is a Senior Product Manager Technical Intern. He loves to build products that help developers learn about various machine learning techniques in a fun and hands-on manner. Phu Nguyen is a Product Manager for AWS DeepLens. He builds products that give developers of any skill level an easy, hands-on introduction to machine learning.
Phu Nguyen is a Product Manager for AWS DeepLens. He builds products that give developers of any skill level an easy, hands-on introduction to machine learning.




















 Julian Bright is an Sr. AI/ML Specialist Solutions Architect based out of Melbourne, Australia. Julian works as part of the global AWS machine learning team and is passionate about helping customers realise their AI and ML journey through MLOps. In his spare time he loves running around after his kids, playing soccer and getting outdoors.
Julian Bright is an Sr. AI/ML Specialist Solutions Architect based out of Melbourne, Australia. Julian works as part of the global AWS machine learning team and is passionate about helping customers realise their AI and ML journey through MLOps. In his spare time he loves running around after his kids, playing soccer and getting outdoors.

 Qingwei Li is a Machine Learning Specialist at Amazon Web Services. He received his Ph.D. in Operations Research after he broke his advisor’s research grant account and failed to deliver the Nobel Prize he promised. Currently he helps customers in financial service and insurance industry build machine learning solutions on AWS. In his spare time, he likes reading and teaching.
Qingwei Li is a Machine Learning Specialist at Amazon Web Services. He received his Ph.D. in Operations Research after he broke his advisor’s research grant account and failed to deliver the Nobel Prize he promised. Currently he helps customers in financial service and insurance industry build machine learning solutions on AWS. In his spare time, he likes reading and teaching. Piali Das is a Senior Software Engineer in the AWS SageMaker Autopilot team. She previously contributed to building SageMaker Algorithms. She enjoys scientific programming in general and has developed an interest in machine learning and distributed systems.
Piali Das is a Senior Software Engineer in the AWS SageMaker Autopilot team. She previously contributed to building SageMaker Algorithms. She enjoys scientific programming in general and has developed an interest in machine learning and distributed systems.

 Muhyun Kim is a data scientist at
Muhyun Kim is a data scientist at 
 Hussain Karimi is a data scientist at the
Hussain Karimi is a data scientist at the 



 Shunjia Ding is a Software Development Engineer working with AWS Step Functions Services development at AWS.
Shunjia Ding is a Software Development Engineer working with AWS Step Functions Services development at AWS.