Creating high-performance machine learning (ML) solutions relies on exploring and optimizing training parameters, also known as hyperparameters. Hyperparameters are the knobs and levers that we use to adjust the training process, such as learning rate, batch size, regularization strength, and others, depending on the specific model and task at hand. Exploring hyperparameters involves systematically varying the values of each parameter and observing the impact on model performance. Although this process requires additional efforts, the benefits are significant. Hyperparameter optimization (HPO) can lead to faster training times, improved model accuracy, and better generalization to new data.
We continue our journey from the post Optimize hyperparameters with Amazon SageMaker Automatic Model Tuning. We previously explored a single job optimization, visualized the outcomes for SageMaker built-in algorithm, and learned about the impact of particular hyperparameter values. On top of using HPO as a one-time optimization at the end of the model creation cycle, we can also use it across multiple steps in a conversational manner. Each tuning job helps us get closer to a good performance, but additionally, we also learn how sensitive the model is to certain hyperparameters and can use this understanding to inform the next tuning job. We can revise the hyperparameters and their value ranges based on what we learned and therefore turn this optimization effort into a conversation. And in the same way that we as ML practitioners accumulate knowledge over these runs, Amazon SageMaker Automatic Model Tuning (AMT) with warm starts can maintain this knowledge acquired in previous tuning jobs for the next tuning job as well.
In this post, we run multiple HPO jobs with a custom training algorithm and different HPO strategies such as Bayesian optimization and random search. We also put warm starts into action and visually compare our trials to refine hyperparameter space exploration.
Advanced concepts of SageMaker AMT
In the next sections, we take a closer look at each of the following topics and show how SageMaker AMT can help you implement them in your ML projects:
- Use custom training code and the popular ML framework Scikit-learn in SageMaker Training
- Define custom evaluation metrics based on the logs for evaluation and optimization
- Perform HPO using an appropriate strategy
- Use warm starts to turn a single hyperparameter search into a dialog with our model
- Use advanced visualization techniques using our solution library to compare two HPO strategies and tuning jobs results
Whether you’re using the built-in algorithms used in our first post or your own training code, SageMaker AMT offers a seamless user experience for optimizing ML models. It provides key functionality that allows you to focus on the ML problem at hand while automatically keeping track of the trials and results. At the same time, it automatically manages the underlying infrastructure for you.
In this post, we move away from a SageMaker built-in algorithm and use custom code. We use a Random Forest from SkLearn. But we stick to the same ML task and dataset as in our first post, which is detecting handwritten digits. We cover the content of the Jupyter notebook 2_advanced_tuning_with_custom_training_and_visualizing.ipynb and invite you to invoke the code side by side to read further.
Let’s dive deeper and discover how we can use custom training code, deploy it, and run it, while exploring the hyperparameter search space to optimize our results.
How to build an ML model and perform hyperparameter optimization
What does a typical process for building an ML solution look like? Although there are many possible use cases and a large variety of ML tasks out there, we suggest the following mental model for a stepwise approach:
- Understand your ML scenario at hand and select an algorithm based on the requirements. For example, you might want to solve an image recognition task using a supervised learning algorithm. In this post, we continue to use the handwritten image recognition scenario and the same dataset as in our first post.
- Decide which implementation of the algorithm in SageMaker Training you want to use. There are various options, inside SageMaker or external ones. Additionally, you need to define which underlying metric fits best for your task and you want to optimize for (such as accuracy, F1 score, or ROC). SageMaker supports four options depending on your needs and resources:
- Use a pre-trained model via Amazon SageMaker JumpStart, which you can use out of the box or just fine-tune it.
- Use one of the built-in algorithms for training and tuning, like XGBoost, as we did in our previous post.
- Train and tune a custom model based on one of the major frameworks like Scikit-learn, TensorFlow, or PyTorch. AWS provides a selection of pre-made Docker images for this purpose. For this post, we use this option, which allows you to experiment quickly by running your own code on top of a pre-made container image.
- Bring your own custom Docker image in case you want to use a framework or software that is not otherwise supported. This option requires the most effort, but also provides the highest degree of flexibility and control.
- Train the model with your data. Depending on the algorithm implementation from the previous step, this can be as simple as referencing your training data and running the training job or by additionally providing custom code for training. In our case, we use some custom training code in Python based on Scikit-learn.
- Apply hyperparameter optimization (as a “conversation” with your ML model). After the training, you typically want to optimize the performance of your model by finding the most promising combination of values for your algorithm’s hyperparameters.
Depending on your ML algorithm and model size, the last step of hyperparameter optimization may turn out to be a bigger challenge than expected. The following questions are typical for ML practitioners at this stage and might sound familiar to you:
- What kind of hyperparameters are impactful for my ML problem?
- How can I effectively search a huge hyperparameter space to find those best-performing values?
- How does the combination of certain hyperparameter values influence my performance metric?
- Costs matter; how can I use my resources in an efficient manner?
- What kind of tuning experiments are worthwhile, and how can I compare them?
It’s not easy to answer these questions, but there is good news. SageMaker AMT takes the heavy lifting from you, and lets you concentrate on choosing the right HPO strategy and value ranges you want to explore. Additionally, our visualization solution facilitates the iterative analysis and experimentation process to efficiently find well-performing hyperparameter values.
In the next sections, we build a digit recognition model from scratch using Scikit-learn and show all these concepts in action.
Solution overview
SageMaker offers some very handy features to train, evaluate, and tune our model. It covers all functionality of an end-to-end ML lifecycle, so we don’t even need to leave our Jupyter notebook.
In our first post, we used the SageMaker built-in algorithm XGBoost. For demonstration purposes, this time we switch to a Random Forest classifier because we can then show how to provide your own training code. We opted for providing our own Python script and using Scikit-learn as our framework. Now, how do we express that we want to use a specific ML framework? As we will see, SageMaker uses another AWS service in the background to retrieve a pre-built Docker container image for training—Amazon Elastic Container Registry (Amazon ECR).
We cover the following steps in detail, including code snippets and diagrams to connect the dots. As mentioned before, if you have the chance, open the notebook and run the code cells step by step to create the artifacts in your AWS environment. There is no better way of active learning.
- First, load and prepare the data. We use Amazon Simple Storage Service (Amazon S3) to upload a file containing our handwritten digits data.
- Next, prepare the training script and framework dependencies. We provide the custom training code in Python, reference some dependent libraries, and make a test run.
- To define the custom objective metrics, SageMaker lets us define a regular expression to extract the metrics we need from the container log files.
- Train the model using the scikit-learn framework. By referencing a pre-built container image, we create a corresponding Estimator object and pass our custom training script.
- AMT enables us to try out various HPO strategies. We concentrate on two of them for this post: random search and Bayesian search.
- Choose between SageMaker HPO strategies.
- Visualize, analyze, and compare tuning results. Our visualization package allows us to discover which strategy performs better and which hyperparameter values deliver the best performance based on our metrics.
- Continue the exploration of the hyperparameter space and warm start HPO jobs.
AMT takes care of scaling and managing the underlying compute infrastructure to run the various tuning jobs on Amazon Elastic Compute Cloud (Amazon EC2) instances. This way, you don’t need to burden yourself to provision instances, handle any operating system and hardware issues, or aggregate log files on your own. The ML framework image is retrieved from Amazon ECR and the model artifacts including tuning results are stored in Amazon S3. All logs and metrics are collected in Amazon CloudWatch for convenient access and further analysis if needed.
Prerequisites
Because this is a continuation of a series, it is recommended, but not necessarily required, to read our first post about SageMaker AMT and HPO. Apart from that, basic familiarity with ML concepts and Python programming is helpful. We also recommend following along with each step in the accompanying notebook from our GitHub repository while reading this post. The notebook can be run independently from the first one, but needs some code from subfolders. Make sure to clone the full repository in your environment as described in the README file.
Experimenting with the code and using the interactive visualization options greatly enhances your learning experience. So, please check it out.
Load and prepare the data
As a first step, we make sure the downloaded digits data we need for training is accessible to SageMaker. Amazon S3 allows us to do this in a safe and scalable way. Refer to the notebook for the complete source code and feel free to adapt it with your own data.
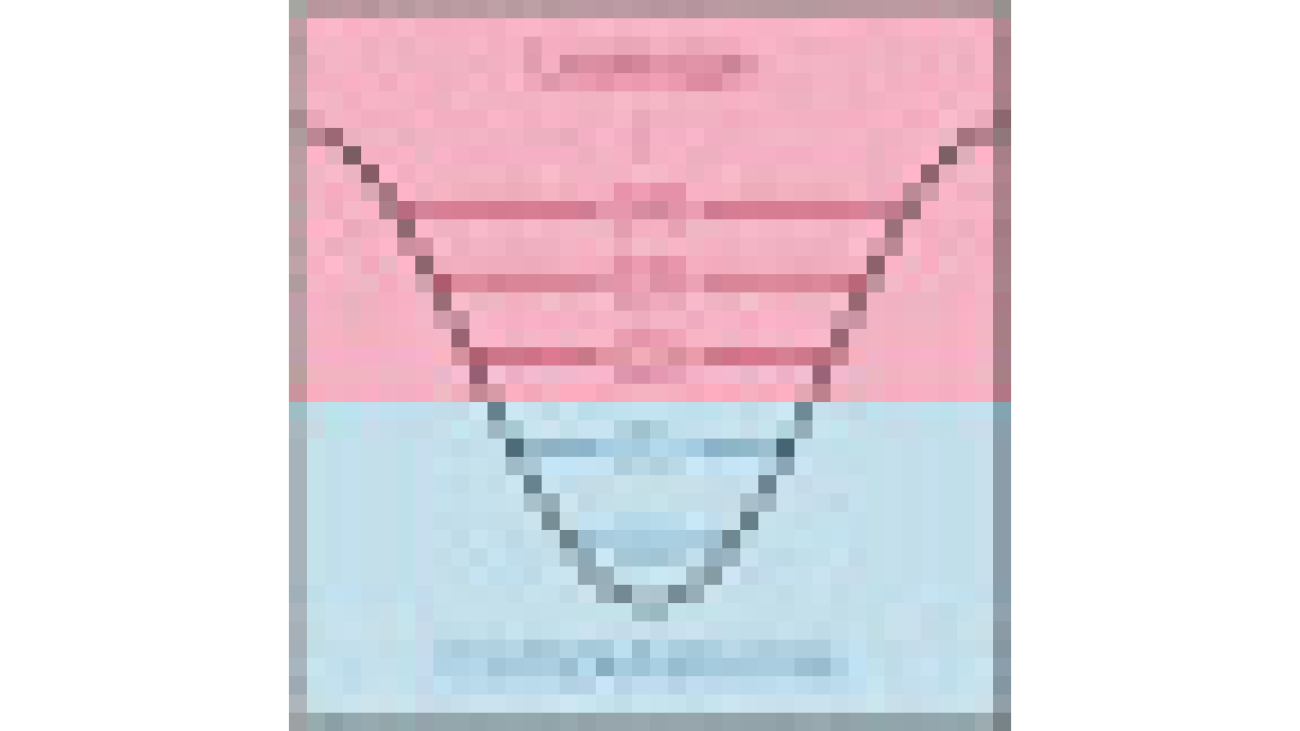
The digits.csv file contains feature data and labels. Each digit is represented by pixel values in an 8×8 image, as depicted by the following image for the digit 4.
Prepare the training script and framework dependencies
Now that the data is stored in our S3 bucket, we can define our custom training script based on Scikit-learn in Python. SageMaker gives us the option to simply reference the Python file later for training. Any dependencies like the Scikit-learn or pandas libraries can be provided in two ways:
- They can be specified explicitly in a
requirements.txtfile - They are pre-installed in the underlying ML container image, which is either provided by SageMaker or custom-built
Both options are generally considered standard ways for dependency management, so you might already be familiar with it. SageMaker supports a variety of ML frameworks in a ready-to-use managed environment. This includes many of the most popular data science and ML frameworks like PyTorch, TensorFlow, or Scikit-learn, as in our case. We don’t use an additional requirements.txt file, but feel free to add some libraries to try it out.
The code of our implementation contains a method called fit(), which creates a new classifier for the digit recognition task and trains it. In contrast to our first post where we used the SageMaker built-in XGBoost algorithm, we now use a RandomForestClassifier provided by the ML library sklearn. The call of the fit() method on the classifier object starts the training process using a subset (80%) of our CSV data:
See the full script in our Jupyter notebook on GitHub.
Before you spin up container resources for the full training process, did you try to run the script directly? This is a good practice to quickly ensure the code has no syntax errors, check for matching dimensions of your data structures, and some other errors early on.
There are two ways to run your code locally. First, you can run it right away in the notebook, which also allows you to use the Python Debugger pdb:
Alternatively, run the train script from the command line in the same way you may want to use it in a container. This also supports setting various parameters and overwriting the default values as needed, for example:
As output, you can see the first results for the model’s performance based on the objective metrics precision, recall, and F1-score. For example, pre: 0.970 rec: 0.969 f1: 0.969.
Not bad for such a quick training. But where did these numbers come from and what do we do with them?
Define custom objective metrics
Remember, our goal is to fully train and tune our model based on the objective metrics we consider relevant for our task. Because we use a custom training script, we need to define those metrics for SageMaker explicitly.
Our script emits the metrics precision, recall, and F1-score during training simply by using the print function:
The standard output is captured by SageMaker and sent to CloudWatch as a log stream. To retrieve the metric values and work with them later in SageMaker AMT, we need to provide some information on how to parse that output. We can achieve this by defining regular expression statements (for more information, refer to Monitor and Analyze Training Jobs Using Amazon CloudWatch Metrics):
Let’s walk through the first metric definition in the preceding code together. SageMaker will look for output in the log that starts with pre: and is followed by one or more whitespace and then a number that we want to extract, which is why we use the round parenthesis. Every time SageMaker finds a value like that, it turns it into a CloudWatch metric with the name valid-precision.
Train the model using the Scikit-learn framework
After we create our training script train.py and instruct SageMaker on how to monitor the metrics within CloudWatch, we define a SageMaker Estimator object. It initiates the training job and uses the instance type we specify. But how can this instance type be different from the one you run an Amazon SageMaker Studio notebook on, and why? SageMaker Studio runs your training (and inference) jobs on separate compute instances than your notebook. This allows you to continue working in your notebook while the jobs run in the background.
The parameter framework_version refers to the Scikit-learn version we use for our training job. Alternatively, we can pass image_uri to the estimator. You can check whether your favorite framework or ML library is available as a pre-built SageMaker Docker image and use it as is or with extensions.
Moreover, we can run SageMaker training jobs on EC2 Spot Instances by setting use_spot_instances to True. They are spare capacity instances that can save up to 90% of costs. These instances provide flexibility on when the training jobs are run.
After the Estimator object is set up, we start the training by calling the fit() function, supplying the path to the training dataset on Amazon S3. We can use this same method to provide validation and test data. We set the wait parameter to True so we can use the trained model in the subsequent code cells.
estimator.fit({'train': s3_data_url}, wait=True)Define hyperparameters and run tuning jobs
So far, we have trained the model with one set of hyperparameter values. But were those values good? Or could we look for better ones? Let’s use the HyperparameterTuner class to run a systematic search over the hyperparameter space. How do we search this space with the tuner? The necessary parameters are the objective metric name and objective type that will guide the optimization. The optimization strategy is another key argument for the tuner because it further defines the search space. The following are four different strategies to choose from:
- Grid search
- Random search
- Bayesian optimization (default)
- Hyperband
We further describe these strategies and equip you with some guidance to choose one later in this post.
Before we define and run our tuner object, let’s recap our understanding from an architecture perspective. We covered the architectural overview of SageMaker AMT in our last post and reproduce an excerpt of it here for convenience.

We can choose what hyperparameters we want to tune or leave static. For dynamic hyperparameters, we provide hyperparameter_ranges that can be used to optimize for tunable hyperparameters. Because we use a Random Forest classifier, we have utilized the hyperparameters from the Scikit-learn Random Forest documentation.
We also limit resources with the maximum number of training jobs and parallel training jobs the tuner can use. We will see how these limits help us compare the results of various strategies with each other.
Similar to the Estimator’s fit function, we start a tuning job calling the tuner’s fit:
This is all we have to do to let SageMaker run the training jobs (n=50) in the background, each using a different set of hyperparameters. We explore the results later in this post. But before that, let’s start another tuning job, this time applying the Bayesian optimization strategy. We will compare both strategies visually after their completion.
Note that both tuner jobs can run in parallel because SageMaker orchestrates the required compute instances independently of each other. That’s quite helpful for practitioners who experiment with different approaches at the same time, like we do here.
Choose between SageMaker HPO strategies
When it comes to tuning strategies, you have a few options with SageMaker AMT: grid search, random search, Bayesian optimization, and Hyperband. These strategies determine how the automatic tuning algorithms explore the specified ranges of hyperparameters.
Random search is pretty straightforward. It randomly selects combinations of values from the specified ranges and can be run in a sequential or parallel manner. It’s like throwing darts blindfolded, hoping to hit the target. We have started with this strategy, but will the results improve with another one?
Bayesian optimization takes a different approach than random search. It considers the history of previous selections and chooses values that are likely to yield the best results. If you want to learn from previous explorations, you can achieve this only with running a new tuning job after the previous ones. Makes sense, right? In this way, Bayesian optimization is dependent on the previous runs. But do you see what HPO strategy allows for higher parallelization?
Hyperband is an interesting one! It uses a multi-fidelity strategy, which means it dynamically allocates resources to the most promising training jobs and stops those that are underperforming. Therefore, Hyperband is computationally efficient with resources, learning from previous training jobs. After stopping the underperforming configurations, a new configuration starts, and its values are chosen randomly.
Depending on your needs and the nature of your model, you can choose between random search, Bayesian optimization, or Hyperband as your tuning strategy. Each has its own approach and advantages, so it’s important to consider which one works best for your ML exploration. The good news for ML practitioners is that you can select the best HPO strategy by visually comparing the impact of each trial on the objective metric. In the next section, we see how to visually identify the impact of different strategies.
Visualize, analyze, and compare tuning results
When our tuning jobs are complete, it gets exciting. What results do they deliver? What kind of boost can you expect on our metric compared to your base model? What are the best-performing hyperparameters for our use case?
A quick and straightforward way to view the HPO results is by visiting the SageMaker console. Under Hyperparameter tuning jobs, we can see (per tuning job) the combination of hyperparameter values that have been tested and delivered the best performance as measured by our objective metric (valid-f1).

Is that all you need? As an ML practitioner, you may be not only interested in those values, but certainly want to learn more about the inner workings of your model to explore its full potential and strengthen your intuition with empirical feedback.
A good visualization tool can greatly help you understand the improvement by HPO over time and get empirical feedback on design decisions of your ML model. It shows the impact of each individual hyperparameter on your objective metric and provides guidance to further optimize your tuning results.
We use the amtviz custom visualization package to visualize and analyze tuning jobs. It’s straightforward to use and provides helpful features. We demonstrate its benefit by interpreting some individual charts, and finally comparing random search side by side with Bayesian optimization.
First, let’s create a visualization for random search. We can do this by calling visualize_tuning_job() from amtviz and passing our first tuner object as an argument:
You will see a couple of charts, but let’s take it step by step. The first scatter plot from the output looks like the following and already gives us some visual clues we wouldn’t recognize in any table.

Each dot represents the performance of an individual training job (our objective valid-f1 on the y-axis) based on its start time (x-axis), produced by a specific set of hyperparameters. Therefore, we look at the performance of our model as it progresses over the duration of the tuning job.
The dotted line highlights the best result found so far and indicates improvement over time. The best two training jobs achieved an F1 score of around 0.91.
Besides the dotted line showing the cumulative progress, do you see a trend in the chart?
Probably not. And this is expected, because we’re viewing the results of the random HPO strategy. Each training job was run using a different but randomly selected set of hyperparameters. If we continued our tuning job (or ran another one with the same setting), we would probably see some better results over time, but we can’t be sure. Randomness is a tricky thing.
The next charts help you gauge the influence of hyperparameters on the overall performance. All hyperparameters are visualized, but for the sake of brevity, we focus on two of them: n-estimators and max-depth.

Our top two training jobs were using n-estimators of around 20 and 80, and max-depth of around 10 and 18, respectively. The exact hyperparameter values are displayed via tooltip for each dot (training job). They are even dynamically highlighted across all charts and give you a multi-dimensional view! Did you see that? Each hyperparameter is plotted against the objective metric, as a separate chart.
Now, what kind of insights do we get about n-estimators?
Based on the left chart, it seems that very low value ranges (below 10) more often deliver poor results compared to higher values. Therefore, higher values may help your model to perform better—interesting.
In contrast, the correlation of the max-depth hyperparameter to our objective metric is rather low. We can’t clearly tell which value ranges are performing better from a general perspective.
In summary, random search can help you find a well-performing set of hyperparameters even in a relatively short amount of time. Also, it isn’t biased towards a good solution but gives a balanced view of the search space. Your resource utilization, however, might not be very efficient. It continues to run training jobs with hyperparameters in value ranges that are known to deliver poor results.
Let’s examine the results of our second tuning job using Bayesian optimization. We can use amtviz to visualize the results in the same way as we did so far for the random search tuner. Or, even better, we can use the capability of the function to compare both tuning jobs in a single set of charts. Quite handy!

There are more dots now because we visualize the results of all training jobs for both, the random search (orange dots) and the Bayesian optimization (blue dots). On the right side, you can see a density chart visualizing the distribution of all F1-scores. A majority of the training jobs achieved results in the upper part of the F1 scale (over 0.6)—that’s good!
What is the key takeaway here? The scatter plot clearly shows the benefit of Bayesian optimization. It delivers better results over time because it can learn from previous runs. That’s why we achieved significantly better results using Bayesian compared to random (0.967 vs. 0.919) with the same number of training jobs.
There is even more you can do with amtviz. Let’s drill in.
If you give SageMaker AMT the instruction to run a larger number of jobs for tuning, seeing many trials at once can get messy. That’s one of the reasons why we made these charts interactive. You can click and drag on every hyperparameter scatter plot to zoom in to certain value ranges and refine your visual interpretation of the results. All other charts are automatically updated. That’s pretty helpful, isn’t it? See the next charts as an example and try it for yourself in your notebook!

As a tuning maximalist, you may also decide that running another hyperparameter tuning job could further improve your model performance. But this time, a more specific range of hyperparameter values can be explored because you already know (roughly) where to expect better results. For example, you may choose to focus on values between 100–200 for n-estimators, as shown in the chart. This lets AMT focus on the most promising training jobs and increases your tuning efficiency.
To sum it up, amtviz provides you with a rich set of visualization capabilities that allow you to better understand the impact of your model’s hyperparameters on performance and enable smarter decisions in your tuning activities.
Continue the exploration of the hyperparameter space and warm start HPO jobs
We have seen that AMT helps us explore the hyperparameter search space efficiently. But what if we need multiple rounds of tuning to iteratively improve our results? As mentioned in the beginning, we want to establish an optimization feedback cycle—our “conversation” with the model. Do we need to start from scratch every time?
Let’s look into the concept of running a warm start hyperparameter tuning job. It doesn’t initiate new tuning jobs from scratch, it reuses what has been learned in the previous HPO runs. This helps us be more efficient with our tuning time and compute resources. We can further iterate on top of our previous results. To use warm starts, we create a WarmStartConfig and specify warm_start_type as IDENTICAL_DATA_AND_ALGORITHM. This means that we change the hyperparameter values but we don’t change the data or algorithm. We tell AMT to transfer the previous knowledge to our new tuning job.
By referring to our previous Bayesian optimization and random search tuning jobs as parents, we can use them both for the warm start:
To see the benefit of using warm starts, refer to the following charts. These are generated by amtviz in a similar way as we did earlier, but this time we have added another tuning job based on a warm start.

In the left chart, we can observe that new tuning jobs mostly lie in the upper-right corner of the performance metric graph (see dots marked in orange). The warm start has indeed reused the previous results, which is why those data points are in the top results for F1 score. This improvement is also reflected in the density chart on the right.
In other words, AMT automatically selects promising sets of hyperparameter values based on its knowledge from previous trials. This is shown in the next chart. For example, the algorithm would test a low value for n-estimators less often because these are known to produce poor F1 scores. We don’t waste any resources on that, thanks to warm starts.

Clean up
To avoid incurring unwanted costs when you’re done experimenting with HPO, you must remove all files in your S3 bucket with the prefix amt-visualize-demo and also shut down SageMaker Studio resources.
Run the following code in your notebook to remove all S3 files from this post:
If you wish to keep the datasets or the model artifacts, you may modify the prefix in the code to amt-visualize-demo/data to only delete the data or amt-visualize-demo/output to only delete the model artifacts.
Conclusion
We have learned how the art of building ML solutions involves exploring and optimizing hyperparameters. Adjusting those knobs and levers is a demanding yet rewarding process that leads to faster training times, improved model accuracy, and overall better ML solutions. The SageMaker AMT functionality helps us run multiple tuning jobs and warm start them, and provides data points for further review, visual comparison, and analysis.
In this post, we looked into HPO strategies that we use with SageMaker AMT. We started with random search, a straightforward but performant strategy where hyperparameters are randomly sampled from a search space. Next, we compared the results to Bayesian optimization, which uses probabilistic models to guide the search for optimal hyperparameters. After we identified a suitable HPO strategy and good hyperparameter value ranges through initial trials, we showed how to use warm starts to streamline future HPO jobs.
You can explore the hyperparameter search space by comparing quantitative results. We have suggested the side-by-side visual comparison and provided the necessary package for interactive exploration. Let us know in the comments how helpful it was for you on your hyperparameter tuning journey!
About the authors
 Ümit Yoldas is a Senior Solutions Architect with Amazon Web Services. He works with enterprise customers across industries in Germany. He’s driven to translate AI concepts into real-world solutions. Outside of work, he enjoys time with family, savoring good food, and pursuing fitness.
Ümit Yoldas is a Senior Solutions Architect with Amazon Web Services. He works with enterprise customers across industries in Germany. He’s driven to translate AI concepts into real-world solutions. Outside of work, he enjoys time with family, savoring good food, and pursuing fitness.
 Elina Lesyk is a Solutions Architect located in Munich. She is focusing on enterprise customers from the financial services industry. In her free time, you can find Elina building applications with generative AI at some IT meetups, driving a new idea on fixing climate change fast, or running in the forest to prepare for a half-marathon with a typical deviation from the planned schedule.
Elina Lesyk is a Solutions Architect located in Munich. She is focusing on enterprise customers from the financial services industry. In her free time, you can find Elina building applications with generative AI at some IT meetups, driving a new idea on fixing climate change fast, or running in the forest to prepare for a half-marathon with a typical deviation from the planned schedule.
 Mariano Kamp is a Principal Solutions Architect with Amazon Web Services. He works with banks and insurance companies in Germany on machine learning. In his spare time, Mariano enjoys hiking with his wife.
Mariano Kamp is a Principal Solutions Architect with Amazon Web Services. He works with banks and insurance companies in Germany on machine learning. In his spare time, Mariano enjoys hiking with his wife.


















































 Gayatri Ghanakota is a Sr. Machine Learning Engineer with AWS Professional Services. She is passionate about developing, deploying, and explaining AI/ ML solutions across various domains. Prior to this role, she led multiple initiatives as a data scientist and ML engineer with top global firms in the financial and retail space. She holds a master’s degree in Computer Science specialized in Data Science from the University of Colorado, Boulder.
Gayatri Ghanakota is a Sr. Machine Learning Engineer with AWS Professional Services. She is passionate about developing, deploying, and explaining AI/ ML solutions across various domains. Prior to this role, she led multiple initiatives as a data scientist and ML engineer with top global firms in the financial and retail space. She holds a master’s degree in Computer Science specialized in Data Science from the University of Colorado, Boulder. Sunita Koppar is a Sr. Data Lake Architect with AWS Professional Services. She is passionate about solving customer pain points processing big data and providing long-term scalable solutions. Prior to this role, she developed products in internet, telecom, and automotive domains, and has been an AWS customer. She holds a master’s degree in Data Science from the University of California, Riverside.
Sunita Koppar is a Sr. Data Lake Architect with AWS Professional Services. She is passionate about solving customer pain points processing big data and providing long-term scalable solutions. Prior to this role, she developed products in internet, telecom, and automotive domains, and has been an AWS customer. She holds a master’s degree in Data Science from the University of California, Riverside. Saswata Dash is a DevOps Consultant with AWS Professional Services. She has worked with customers across healthcare and life sciences, aviation, and manufacturing. She is passionate about all things automation and has comprehensive experience in designing and building enterprise-scale customer solutions in AWS. Outside of work, she pursues her passion for photography and catching sunrises.
Saswata Dash is a DevOps Consultant with AWS Professional Services. She has worked with customers across healthcare and life sciences, aviation, and manufacturing. She is passionate about all things automation and has comprehensive experience in designing and building enterprise-scale customer solutions in AWS. Outside of work, she pursues her passion for photography and catching sunrises.

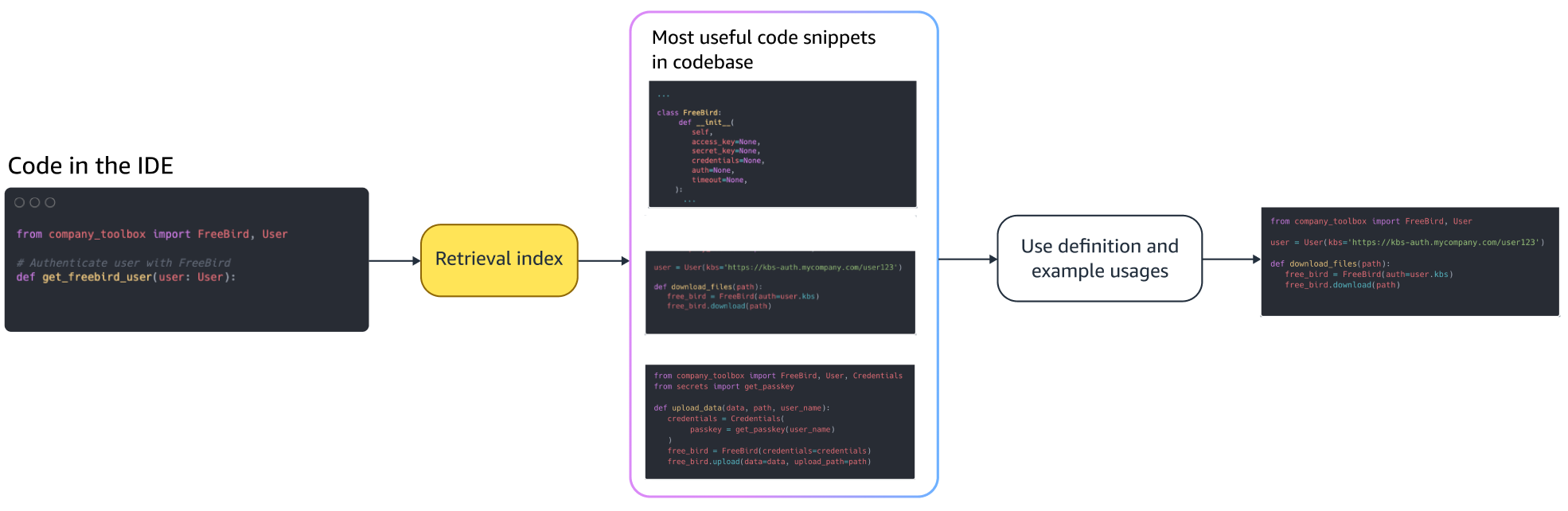




 Qing Sun is a Senior Applied Scientist in AWS AI Labs and work on AWS CodeWhisperer, a generative AI-powered coding assistant. Her research interests lie in Natural Language Processing, AI4Code and generative AI. In the past, she had worked on several NLP-based services such as Comprehend Medical, a medical diagnosis system at Amazon Health AI and Machine Translation system at Meta AI. She received her PhD from Virginia Tech in 2017.
Qing Sun is a Senior Applied Scientist in AWS AI Labs and work on AWS CodeWhisperer, a generative AI-powered coding assistant. Her research interests lie in Natural Language Processing, AI4Code and generative AI. In the past, she had worked on several NLP-based services such as Comprehend Medical, a medical diagnosis system at Amazon Health AI and Machine Translation system at Meta AI. She received her PhD from Virginia Tech in 2017. Arash Farahani is an Applied Scientist with Amazon CodeWhisperer. His current interests are in generative AI, search, and personalization. Arash is passionate about building solutions that resolve developer pain points. He has worked on multiple features within CodeWhisperer, and introduced NLP solutions into various internal workstreams that touch all Amazon developers. He received his PhD from University of Illinois at Urbana-Champaign in 2017.
Arash Farahani is an Applied Scientist with Amazon CodeWhisperer. His current interests are in generative AI, search, and personalization. Arash is passionate about building solutions that resolve developer pain points. He has worked on multiple features within CodeWhisperer, and introduced NLP solutions into various internal workstreams that touch all Amazon developers. He received his PhD from University of Illinois at Urbana-Champaign in 2017. Xiaofei Ma is an Applied Science Manager in AWS AI Labs. He joined Amazon in 2016 as an Applied Scientist within SCOT organization and then later AWS AI Labs in 2018 working on Amazon Kendra. Xiaofei has been serving as the science manager for several services including Kendra, Contact Lens, and most recently CodeWhisperer and CodeGuru Security. His research interests lie in the area of AI4Code and Natural Language Processing. He received his PhD from University of Maryland, College Park in 2010.
Xiaofei Ma is an Applied Science Manager in AWS AI Labs. He joined Amazon in 2016 as an Applied Scientist within SCOT organization and then later AWS AI Labs in 2018 working on Amazon Kendra. Xiaofei has been serving as the science manager for several services including Kendra, Contact Lens, and most recently CodeWhisperer and CodeGuru Security. His research interests lie in the area of AI4Code and Natural Language Processing. He received his PhD from University of Maryland, College Park in 2010. Murali Krishna Ramanathan is a Principal Applied Scientist in AWS AI Labs and co-leads AWS CodeWhisperer, a generative AI-powered coding companion. He is passionate about building software tools and workflows that help improve developer productivity. In the past, he built Piranha, an automated refactoring tool to delete code due to stale feature flags and led code quality initiatives at Uber engineering. He is a recipient of the Google faculty award (2015), ACM SIGSOFT Distinguished paper award (ISSTA 2016) and Maurice Halstead award (Purdue 2006). He received his PhD in Computer Science from Purdue University in 2008.
Murali Krishna Ramanathan is a Principal Applied Scientist in AWS AI Labs and co-leads AWS CodeWhisperer, a generative AI-powered coding companion. He is passionate about building software tools and workflows that help improve developer productivity. In the past, he built Piranha, an automated refactoring tool to delete code due to stale feature flags and led code quality initiatives at Uber engineering. He is a recipient of the Google faculty award (2015), ACM SIGSOFT Distinguished paper award (ISSTA 2016) and Maurice Halstead award (Purdue 2006). He received his PhD in Computer Science from Purdue University in 2008. Ramesh Nallapati is a Senior Principal Applied Scientist in AWS AI Labs and co-leads CodeWhisperer, a generative AI-powered coding companion, and Titan Large Language Models at AWS. His interests are mainly in the areas of Natural Language Processing and Generative AI. In the past, Ramesh has provided science leadership in delivering many NLP-based AWS products such as Kendra, Quicksight Q and Contact Lens. He held research positions at Stanford, CMU and IBM Research, and received his Ph.D. in Computer Science from University of Massachusetts Amherst in 2006.
Ramesh Nallapati is a Senior Principal Applied Scientist in AWS AI Labs and co-leads CodeWhisperer, a generative AI-powered coding companion, and Titan Large Language Models at AWS. His interests are mainly in the areas of Natural Language Processing and Generative AI. In the past, Ramesh has provided science leadership in delivering many NLP-based AWS products such as Kendra, Quicksight Q and Contact Lens. He held research positions at Stanford, CMU and IBM Research, and received his Ph.D. in Computer Science from University of Massachusetts Amherst in 2006.



 NVIDIA GeForce NOW (@NVIDIAGFN)
NVIDIA GeForce NOW (@NVIDIAGFN) 


 David Bericat is a product manager for Healthcare at NVIDIA, where he leads the Project MONAI Deploy working group to bring AI from research to clinical deployments. His passion is to accelerate health innovation globally translating it to true clinical impact. Previously, David worked at Red Hat, implementing open source principles at the intersection of AI, cloud, edge computing, and IoT. His proudest moments include hiking to the Everest base camp and playing soccer for over 20 years.
David Bericat is a product manager for Healthcare at NVIDIA, where he leads the Project MONAI Deploy working group to bring AI from research to clinical deployments. His passion is to accelerate health innovation globally translating it to true clinical impact. Previously, David worked at Red Hat, implementing open source principles at the intersection of AI, cloud, edge computing, and IoT. His proudest moments include hiking to the Everest base camp and playing soccer for over 20 years. Brad Genereaux is Global Lead, Healthcare Alliances at NVIDIA, where he is responsible for developer relations with a focus in medical imaging to accelerate artificial intelligence and deep learning, visualization, virtualization, and analytics solutions. Brad evangelizes the ubiquitous adoption and integration of seamless healthcare and medical imaging workflows into everyday clinical practice, with more than 20 years of experience in healthcare IT.
Brad Genereaux is Global Lead, Healthcare Alliances at NVIDIA, where he is responsible for developer relations with a focus in medical imaging to accelerate artificial intelligence and deep learning, visualization, virtualization, and analytics solutions. Brad evangelizes the ubiquitous adoption and integration of seamless healthcare and medical imaging workflows into everyday clinical practice, with more than 20 years of experience in healthcare IT. Gang Fu is a Healthcare Solutions Architect at AWS. He holds a PhD in Pharmaceutical Science from the University of Mississippi and has over 10 years of technology and biomedical research experience. He is passionate about technology and the impact it can make on healthcare.
Gang Fu is a Healthcare Solutions Architect at AWS. He holds a PhD in Pharmaceutical Science from the University of Mississippi and has over 10 years of technology and biomedical research experience. He is passionate about technology and the impact it can make on healthcare. JP Leger is a Senior Solutions Architect supporting academic medical centers and medical imaging workflows at AWS. He has over 20 years of expertise in software engineering, healthcare IT, and medical imaging, with extensive experience architecting systems for performance, scalability, and security in distributed deployments of large data volumes on premises, in the cloud, and hybrid with analytics and AI.
JP Leger is a Senior Solutions Architect supporting academic medical centers and medical imaging workflows at AWS. He has over 20 years of expertise in software engineering, healthcare IT, and medical imaging, with extensive experience architecting systems for performance, scalability, and security in distributed deployments of large data volumes on premises, in the cloud, and hybrid with analytics and AI. Chris Hafey is a Principal Solutions Architect at Amazon Web Services. He has over 25 years’ experience in the medical imaging industry and specializes in building scalable high-performance systems. He is the creator of the popular CornerstoneJS open source project, which powers the popular OHIF open source zero footprint viewer. He contributed to the DICOMweb specification and continues to work towards improving its performance for web-based viewing.
Chris Hafey is a Principal Solutions Architect at Amazon Web Services. He has over 25 years’ experience in the medical imaging industry and specializes in building scalable high-performance systems. He is the creator of the popular CornerstoneJS open source project, which powers the popular OHIF open source zero footprint viewer. He contributed to the DICOMweb specification and continues to work towards improving its performance for web-based viewing.